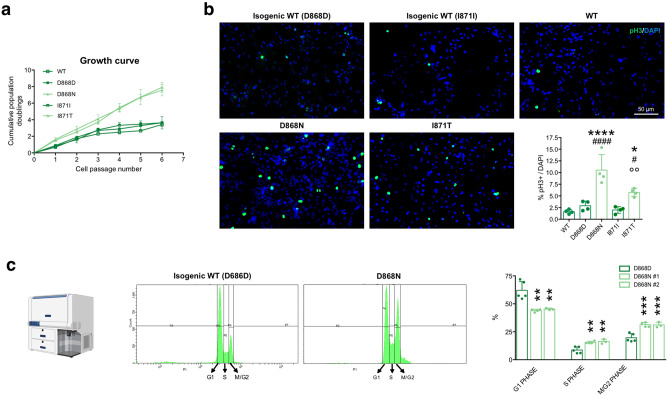
Fig. 3. SGS NPCs display overproliferation.
a Growth curve analysis of WT, isogenic control, and mutant NPCs See Supplementary Dataset 3 for statistical analysis. n = 3. b NPC proliferation assay by phosphorylated H3 (pH3 Ser10) immunostaining and quantification in WT, isogenic control, and mutant NPCs. * indicates statistic test between isogenic cell line pairs, # refers to the comparison between each mutant cell line with the unrelated WT one, ° refers to the comparison between mutant NPCs. P < 0.0001; WT vs. D868D P = 0.8012; WT vs. D868N ####P < 0.0001; WT vs. I871I P = 0.9972; WT vs. I871T #P = 0.0202; D868D vs. D868N ****P < 0.0001; I871I vs. I871T *P = 0.0377; D868N vs. I871T °°P = 0.0068. See Supplementary Fig. 2f for WT vs. MUT comparisons. n = 4. Images taken at the same magnification. c Cell cycle analysis by PI staining and FACS quantification; graphs showing different peak distribution indicating different cell cycle phases in isogenic control and mutant NPCs. Bar graph shows the percentage of cells present in each phase in D868D and D868N cells. G1: D868D vs. D868N #1 ****P = <0.0001, D868D vs. D868N #2 ****P < 0.0001; S: D868D vs. D868N #1 *P = 0.0311, D868D vs. D868N #2 *P = 0.0253; M-G2: D868D vs. D868N #1 ***P = 0.0002, D868D vs. D868N #2 ***P = 0.0007. n = 3 independent experiments. All data expressed as mean ± SEM. ns: nonsignificant differences when P > 0.05; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. a, c Two-way ANOVA followed by Tukey post hoc test for multiple comparisons, b one-way ANOVA followed by Tukey post hoc test for multiple comparisons.