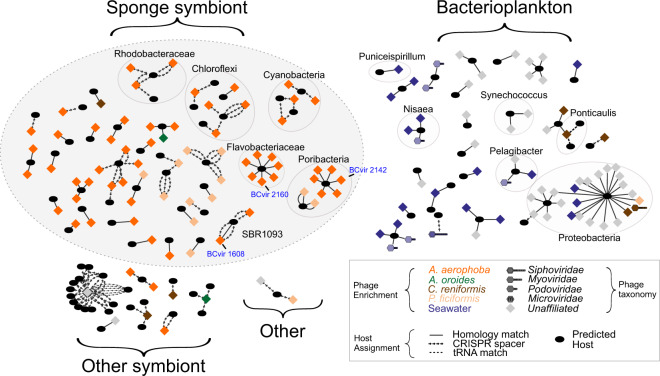
Fig. 1. Predicted phage–bacteria infection networks.
Phage (diamond) assignments to putative bacterial hosts (oval) by combining signals from CRISPR-spacer-, homology- and tRNA matches against a custom database of sponge microbial sequences, PATRIC genomes [81] and 290 Tara Oceans bins from bacterioplankton of the Mediterranean Sea (TMED; Tully et al. [39]). Nodes are organised in groups by niche of the predicted host. ‘Other symbionts’ were isolated from eukaryote hosts including fish and sea cucumber. Phage taxonomy was assigned using reticulate classification based on gene sharing with ViralRefseq entries as performed in Jahn et al. [12]. The symbiont phages subjected to imaging in the next section are labelled in blue text. BCvir 2312 and adjacent nodes were excluded from this representation to aid visualisation. A full network is provided in Supplementary Fig. S4; the underlying raw data are provided in Supplementary Table S5.