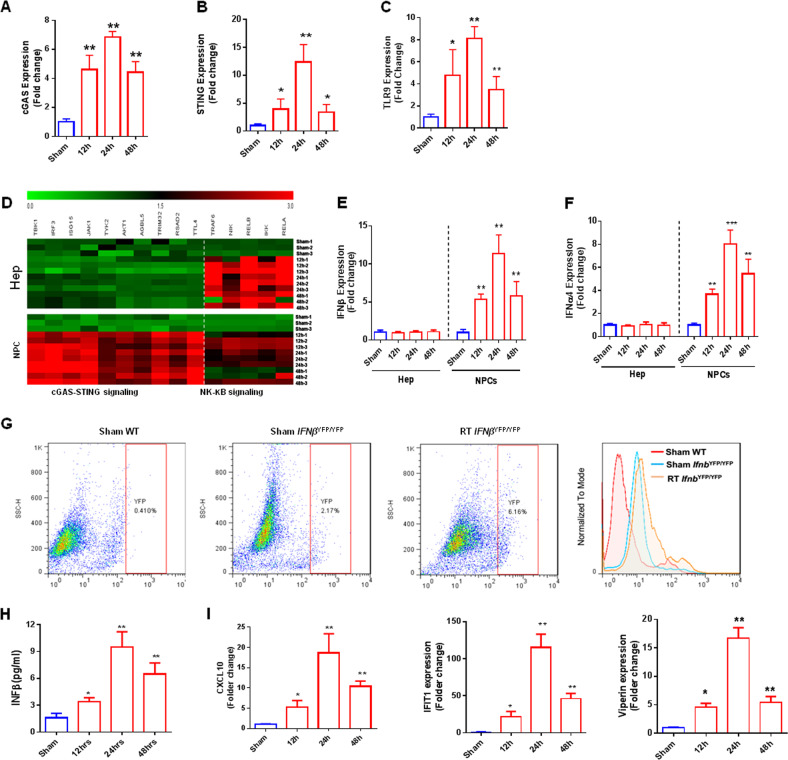
Fig. 3.
Evaluation of DNA sensor gene expression in different hepatic populations after radiation-induced injury. Gene expression of various DNA sensors in liver NPCs and hepatocytes after hepatic irradiation: a cGAS, b STING, and c TLR9. d Heatmap of cGAS-SING-related genes (TBK1, IRF3, ISG15, JAK1, TYK2, AKT1, AGBL5, TRIM32, RSAD2, TTL4) and NF-κB-related genes (TRAF6, NIK, RELB, IKK, RELA) at different time points after radiation-induced hepatic injury. Green = decreased expression; black = no variation; red = increased expression. The graphs represent the mean expressions of selected genes relative to sham-IR hepatocytes (▵▵CT). Gene expression of different cytokines in hepatocytes and NPCs after hepatic irradiation: e IFNβ and f IFNa4. g Flow cytometry for evaluating type 1 IFN production by NPCs in IFNβYFP/YFP mice. h Levels of type 1 IFNs in hepatic TIF. i Evaluation of type 1 IFN-regulated gene expression in hepatocytes after induction of hepatic radiation injury. Mean ± SD; n = 5. *P ≤ 0.05 and ***P ≤ 0.001 compared with control hepatocytes