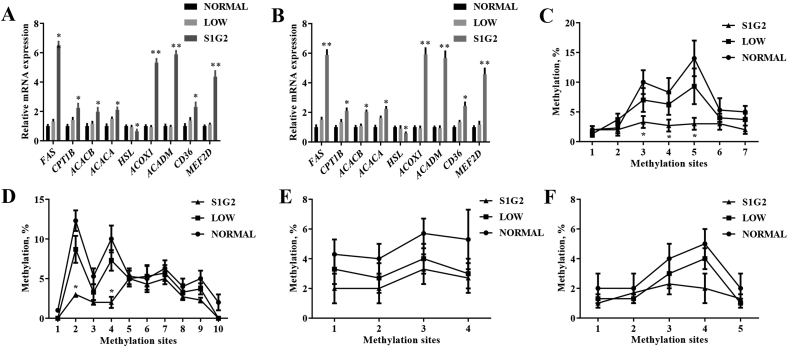
Fig. 3.
Differentially expressed genes involved in lipid metabolism and status of promoter DNA methylation. (A) Relative expression of differentially expressed genes according to RNA-seq sequencing (n = 3). (B) Verification of transcriptomics quantification by qRT-PCR (n = 8). The methylation status of representative CpG sites from promoter sequence of (C) ACOX1, (D) ACADM, (E) FAS and (F) MEF2D genes, respectively (n = 3). NORMAL, pigs fed a normal CP (16%) diet; LOW, pigs fed a low CP (12%) diet generated by decreasing the content of soybean meal; S1G2, pigs fed a low CP (12%) diet with serine-to-glycine ratio 1:2, generated by decreasing the content of soybean meal and adding 0.57% glycine. CP = crude protein. FAS = Fatty acid synthase; CPT1B = Carnitine palmityl transferase 1B; ACACB = acetyl-CoA carboxylase beta; ACACA = acetyl-CoA carboxylase alpha; HSL = lipase E, hormone sensitive type; ACOX1 = acyl-CoA oxidase 1; ACADM = acyl-CoA dehydrogenase medium chain; MEF2D = myocyte enhancer factor 2D. Values are expressed as means ± standard deviations. Mean values were significantly different among groups (∗P < 0.05, ∗∗P < 0.01).