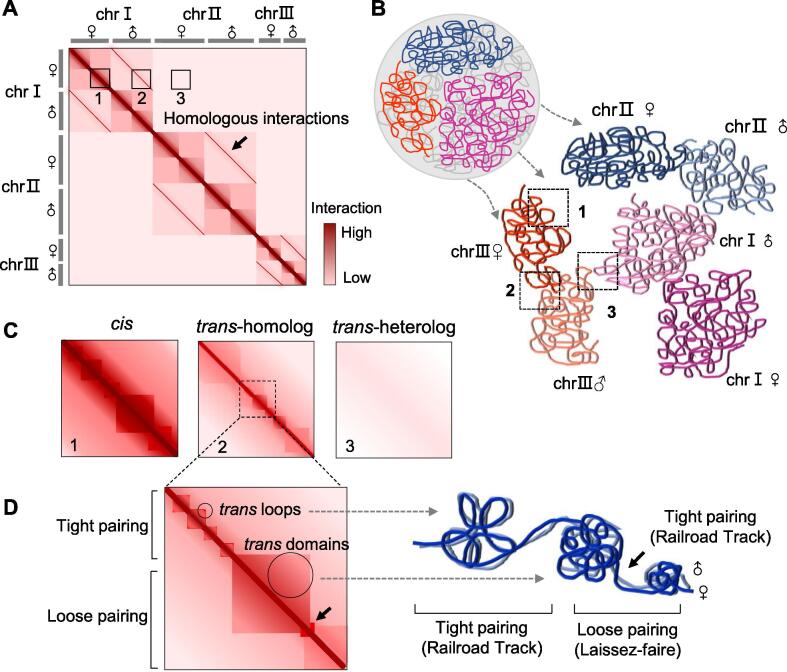
Fig. 2.
Schematics of highly structured homolog pairing in diploid genomes. (A) Haplotype-resolved Hi-C map of a diploid F1 hybrid, which is assumed to have three pairs of chromosomes. The three boxes on the map show the chromatin interactions within the haploid chromosome (cis contacts, box 1), between homologous chromosomes (trans homolog contacts, box 2), and between heterologous chromosomes (trans heterolog contacts, box 3). The t-homo diagonals along the main cis diagonal of the map indicate the homolog pairing. (B) Spatial organization of haploid-level chromosomes in the 3D nucleus of diploid F1 hybrids obtained by 3D modeling of the haploid-resolved Hi-C map. Cis (box 1), t-homo (box 2), and t-heter (box 3) contacts are indicated on the map. (C) Zoomed in Hi-C maps of boxes 1, 2, and 3 shown in (A). (D) Local contact map and 3D organization of variable homolog pairing. Homolog pairing encompasses tightly and loosely paired regions, and displays highly structured trans-domains and trans-interaction peaks.