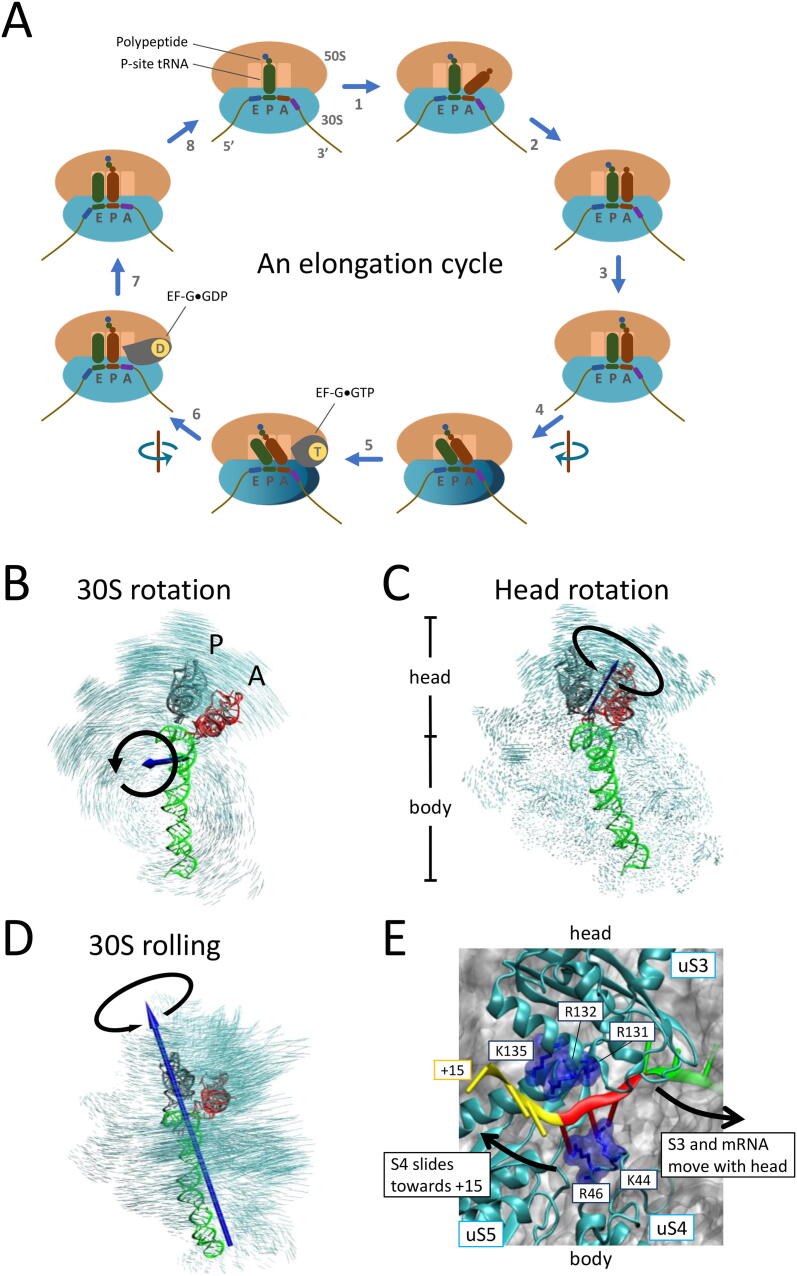
Fig. 2.
Conformational changes of the bacterial ribosome during translation elongation. (A) Cycle of elongation. For clarity, only the aa-tRNA (red; instead of the ternary complex) is shown in step 1, and the swiveling of the 30S head is not shown in steps 5–7. See the text for details. (B) 30S subunit rotation during translocation. Calculated by superimposing the 50S subunit (not shown). (C) 30S head rotation (swiveling). Calculated by superimposing the 30S body. Rotational axes are calculated by pseudo-angular momentum, and the non-rotated and rotated states of the ribosome are modeled as described [38]. (D) 30S subunit rolling during pseudoknot-induced −1 PRF [38]. The 50S subunit (not shown) is superimposed. The rolling axis lies approximately along helix 44 (green ribbon). (B)–(D) are viewed from the solvent side. (E) mRNA entrance site of the 30S subunit (PDB ID: 6BY1) [67]. uS3 is on the 30S head, while uS4 and uS5 are on the 30S body. R131, R132, and K135 of uS3, as well as K44 and R46 of uS4, are highlighted in blue. The relative movement of the 30S head and body described in (C) are indicated with black arrows. The codons at positions +13 to +15, +10 to +12, and +7 to +9 are shown in yellow, red, and green ribbons, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)