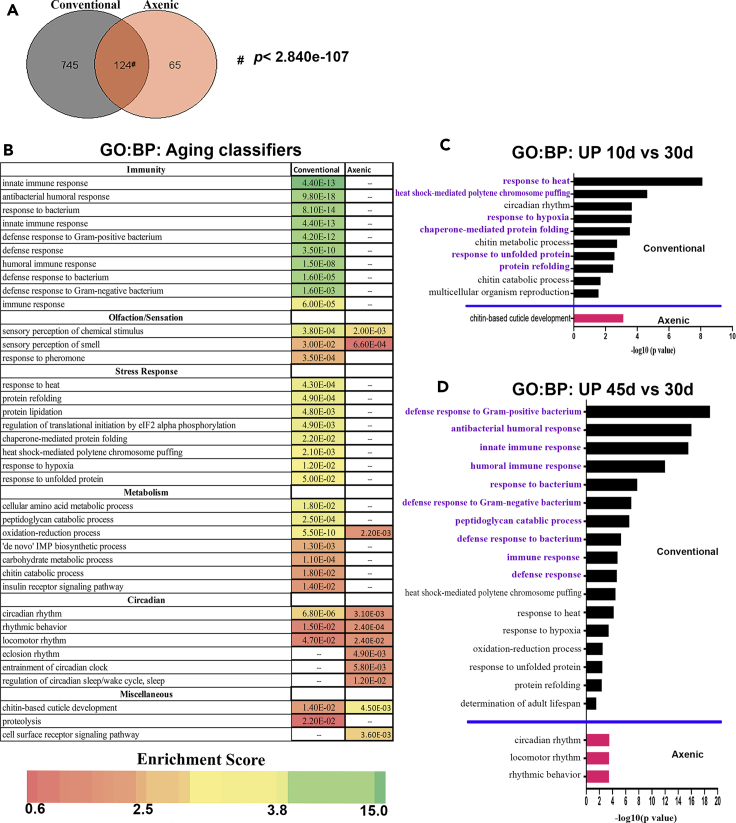
Figure 4.
Gene ontology analysis of aging classifier genes and comparison of conventional vs axenic conditions
(A) Venn diagram showing the number of aging classifiers common between conventional flies (black circle) and axenic (orange circle). Significance assessed by a hypergeometric distribution test: http://nemates.org/MA/progs/overlap_stats.html. For this figure, classifiers for the conventional condition were selected based only on data from 10-, 30-, and 45-day data to facilitate comparison to the axenic data.
(B) Comparison of significantly enriched gene ontology (GO) groups for biological processes (BP) in aging classifier gene lists from conventionally-raised vs axenic-raised flies. Full list of classifier genes is in Table S8, and genes that belong to individual biological processes is in Table S9. Heatmap is based on DAVID enrichment scores and value in each cell is the pvalue calculated by DAVID; cells marked with ‘---’ were not enriched)
(C) Significantly enriched (p≤0.05), biological process terms for genes upregulated in 10-day-old conventional (black bars) and axenic (red bar) raised flies, each compared to its respective 30-day-old condition. Biological processes related to stress response are highlighted in purple. Full list of genes used for analysis is in Table S10 and genes that belong to individual biological processes is in Table S11.
(D) Significantly enriched (p ≤ 0.05) biological process terms for genes upregulated in 45-day-old conventional (black bars) and axenic (red bars) raised flies, each compared to its respective 30-day-old condition. Biological processes related to immunity are highlighted in purple. Full list of genes used for analysis is in Table S10 and genes that belong to individual biological processes is in Table S11.
See also Figure S7.