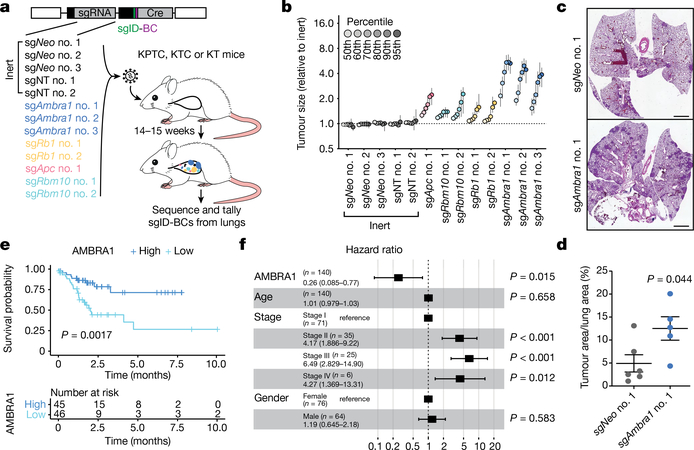
Fig. 4 |. AMBRA1 is a tumour suppressor in KRAS-mutant lung adenocarcinoma.
a, Schematic of multiplexed CRISPR–Cas9 gene editing and Tuba-seq in KPTC mice, KTC mice (wild type for p53) and KT mice (wild type for p53 and lacking Cas9). sgID-BC, dual barcode to identify each individual tumour and its associated sgRNA. Neo denotes the neomycin resistance gene. b, Relative tumour sizes for each sgRNA in KTC mice (n = 9 mice). Tumour sizes were calculated from merged data from all tumours in all mice and normalized to inert sgRNAs 15 weeks after cancer initiation. Error bars denote 95% confidence intervals determined by bootstrap sampling. c, Representative haematoxylin and eosin (H&E) staining of lung sections from control and Ambra1-mutant KC mice 15 weeks after cancer initiation. Scale bars, 1 mm. d, Quantification of tumours in c. n = 6 (sgNeo no. 1) or 5 (sgAmbra1 no. 1) mice. Data are mean ± s.e.m. e, Kaplan–Meier plot of AMBRA1 expression (high, upper third; low, bottom third) in TCGA KRAS G12-mutant lung adenocarcinoma (n = 136 patients). f, Forest plot of Cox proportional hazard model of TCGA KRAS G12-mutant lung adenocarcinoma (n = 131 patients). Model is adjusted by stage, age and gender. Hazard ratios are given with 95% confidence interval in parentheses. P values calculated by two-sided unpaired t-test (d), log-rank test (e) and Wald test (f).