Summary
Defects in protein quality control are the underlying cause of age-related diseases. The western blot analysis of detergent-soluble and insoluble protein fractions has proven useful in identifying interventions that regulate proteostasis. Here, we describe the protocol for such analyses in Drosophila tissues, mouse skeletal muscle, human organoids, and HEK293 cells. We describe key adaptations of this protocol and provide key information that will help modify this protocol for future studies in other tissues and disease models.
For complete details on the use and execution of this protocol, please refer to Rai et al. (2021) and Hunt el al. (2021).
Subject areas: Cell Biology, Developmental biology, Model Organisms, Molecular Biology, Protein Biochemistry, Organoids
Graphical abstract
Highlights
-
•
Testing detergent-soluble and insoluble protein fractions estimates proteostasis
-
•
Loss of proteostasis increases detergent-insoluble poly-ubiquitinated proteins
-
•
Protocol for Drosophila, mouse skeletal muscle, human organoids, and cells
-
•
Protocol can be adapted to other tissues and disease models
Defects in protein quality control are the underlying cause of age-related disease. The western blot analysis of detergent-soluble and insoluble protein fractions has proven useful in identifying interventions that regulate proteostasis. Here, we describe the protocol for such analyses in Drosophila tissues, mouse skeletal muscle, human organoids, and HEK293 cells. We describe key adaptations of this protocol and provide key information that will help modify this protocol for future studies in other tissues and disease models.
Before you begin
Proteostasis decline with aging and defects in protein quality control are the underlying cause of multiple age-related diseases (Douglas and Dillin, 2010; Labbadia and Morimoto, 2015). We have recently found that proteostasis declines during skeletal muscle aging (sarcopenia) in mice (Demontis et al., 2013; Hunt et al., 2021; Jiao and Demontis, 2017), and that protein quality control is compromised across tissues and organ systems in the fruit fly Drosophila during aging (Demontis and Perrimon, 2010; Rai et al., 2021). Moreover, by using thermal stress as a proxy for age-induced challenge to proteostasis, we have investigated the role of genetic and chemical interventions on proteostasis of human brain organoids and HEK293 cells (Rai et al., 2021). Below, we describe the protocols for the analysis of detergent-soluble and insoluble protein fractions in these models and provide details that can help adapt this protocol for the analysis of proteostasis in other experimental systems.
Generation of tissues and cells for analyses
Tissues from Drosophila and mice of the desired genotypes and ages, and treatment of HKE293 cells and human brain organoids should be done according to the specific experimental requirements and following procedures as described in our and other recent studies (Hunt et al., 2019a; Hunt et al., 2021; Hunt et al., 2019b; Hunt et al., 2015; Rai et al., 2021). Once obtained, these tissues can be examined following the procedures described below. Because the necessary buffers are freshly made, no stock solutions are kept.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-poly-ubiquitin (FK2) | Enzo Life Sciences | BML-PW8810-0100 |
| Rabbit anti-Ref(2)P/p62 | Abcam | 178840 |
| Rabbit anti-p62/SQSTM1 | Cell Signaling Technology | 5114 |
| Mouse anti-ubiquitin (P4D1) | Santa Cruz | Sc-8017 |
| Rabbit anti-alpha-tubulin (11H10) | Cell Signaling Technology | 2125S |
| Rabbit anti-Atg8/GABARAP | Abcam | Ab109364 |
| Rabbit anti-beta-actin | Cell Signaling Technology | 8457 |
| Rabbit anti-GAPDH (14C10) | Cell Signaling Technology | 2118 |
| Rabbit anti-Histone H3 (D1H2) | Cell Signaling Technology | 4499 |
| Anti-mouse IgG, HRP-linked | Cell Signaling Technology | 7076S |
| Anti-rabbit IgG, HRP-linked | Cell Signaling Technology | 7074S |
| Chemicals, peptides, and recombinant proteins | ||
| PBS | Gibco | 10010023 |
| RIPA buffer (10×) | Cell Signaling Technology | 9806 |
| NP40 cell lysis buffer | Invitrogen | FNN0021 |
| Protease inhibitors | Roche | 11836153001 |
| Phosphatase inhibitors | Sigma | P5726 |
| 0.5-mm Zirconium oxide beads | NextAdvance | ZROB05 |
| 1.5 mL NAVY Bead Lysis Kit | NextAdvance | NAVY5E100 |
| Blue Loading Buffer Pack | Cell Signaling Technology | 7722 |
| Precision Plus Protein Standard | Bio-Rad | 1610374 |
| 4%–20% Mini-PROTEAN TGX pre-cast gels | Bio-Rad | 4561096 |
| Urea | Sigma-Aldrich | U5378 |
| TritonX-100 | Sigma-Aldrich | T8787 |
| SDS | Sigma-Aldrich | L3771 |
| Immobilon-P PVDF membrane | Millipore | IPVH00010 |
| Ponceau S | Sigma-Aldrich | P7170-1L |
| Benzonase nuclease | Sigma-Aldrich | E1014 |
| Critical commercial assays | ||
| Pierce BCA Protein Assay Kit | Thermo Scientific | 23225 |
| Experimental models: organisms/strains | ||
| Human: HEK293T cells | ATCC | CRL-3216 |
| Drosophila melanogaster | Multiple sources | multiple stocks |
| Mouse | Multiple sources | multiple strains |
| Software and algorithms | ||
| Photoshop CSX | Adobe | https://www.adobe.com/products/photoshop.html |
| GraphPad Prism | Graphpad | https://www.graphpad.com/ |
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
Materials and equipment
Equipment: This protocol requires common lab supplies and equipment, including a refrigerated centrifuge, a tissue homogenizer, and pipettors. Most of the steps should be performed in a cold room (4°C) to limit protein denaturation. In particular, because tissue homogenization can raise the sample temperature, the tissue homogenizer should be used in a cold room (4°C).
Data Analysis Program: Image acquisition and presentation is done with Photoshop (Adobe) and band intensity can be examined with the Histogram function of Photoshop or with ImageJ. Estimated intensities can be further analyzed with Excel or GraphPad Prism.
CRITICAL: Several chemicals used in this protocol are toxic. Before starting, the corresponding safety data sheets should be consulted, and personal protective equipment worn.
Triton X-100 cell lysis buffer (for use with Drosophila, organoids, HEK293 cells)
| Reagent | Final concentration | Amount |
|---|---|---|
| TritonX-100 | 1% | 100 μL |
| phosphatase inhibitors | 1× | 100 μL |
| protease inhibitors | 1× | 1 tablet |
| PBS | bring to 10 mL |
Prepare fresh and keep on ice; discard after use.
Urea and SDS solubilization buffer(for use with Drosophila tissues and organoids)
| Reagent | Final concentration | Amount |
|---|---|---|
| Urea | 8M Urea | 4.8 g |
| 20% SDS | 5% SDS | 2.5 mL |
| 10× RIPA buffer | 1× | 1 mL |
| phosphatase inhibitors | 1× | 100 μL |
| protease inhibitors | 1× | 1 tablet |
| ddH2O | bring to 10 mL |
Prepare fresh and keep at 22°C–25°C for use..
Urea buffer (for HEK293 cells)
| Reagent | Final concentration | Amount |
|---|---|---|
| Urea | 7M Urea | 4.2 g |
| 10× RIPA buffer | 1× | 1 mL |
| phosphatase inhibitors | 1× | 100 μL |
| protease inhibitors | 1× | 1 tablet |
| ddH2O | bring to 10 mL |
Prepare fresh and keep at 22°C–25°C for use.
Urea and SDS solubilization buffer (for mouse skeletal muscle)
| Reagent | Final concentration | Amount |
|---|---|---|
| Urea | 8M | 4.8 g |
| 20% SDS | 1% | 0.5 mL |
| phosphatase inhibitors | 1× | 100 μL |
| protease inhibitors | 1× | 1 tablet |
| PBS | bring to 10 mL |
Prepare fresh and keep at 22°C–25°C for use.
| Solutions | |
|---|---|
| Name | Reagent |
| 20% SDS solution | 20 g SDS in 100 mL of distilled water. |
Store at 22°C–25°C for 2–3 months.
Step-by-step method details
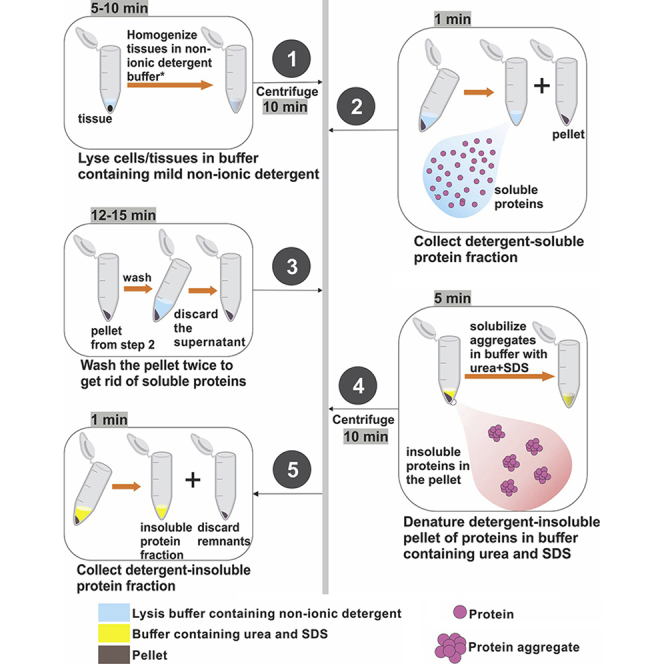
Preparation of detergent-soluble and detergent-insoluble fractions from Drosophila tissues
Timing: 1–2 h (depending on the number of samples)
Western blots of detergent-soluble and insoluble fractions can be obtained substantially as described previously (Demontis and Perrimon, 2010) with the following procedures.
-
1.Homogenize Drosophila tissues in Triton X-100 lysis buffer with the aid of a tissue homogenizer.
-
a.Thoraces, heads, or specific fly tissues are dissected from 30 or more flies (corresponding to around >3 mg of tissue) and homogenized in 90 μL (3 μL/tissue) of ice-cold Triton X-100 buffer, i.e., 1% Triton X-100 in PBS that contains EDTA and that is MgCl2- and CaCl2-free (Gibco #10010023) with protease inhibitors (1 tablet of protease inhibitor cocktail mix in 10 mL of buffer; Roche #11836153001) and phosphatase inhibitors (100 μL of phosphatase inhibitor cocktail mix in 10 mL of buffer, Sigma #P5726).
-
b.The homogenization is done in a tissue homogenizer (NextAdvance) with 0.5-mm zirconium oxide beads at 4°C at maximum speed for 5 min.
-
c.All samples should be homogenized in safe-lock microcentrifuge tubes.
-
a.
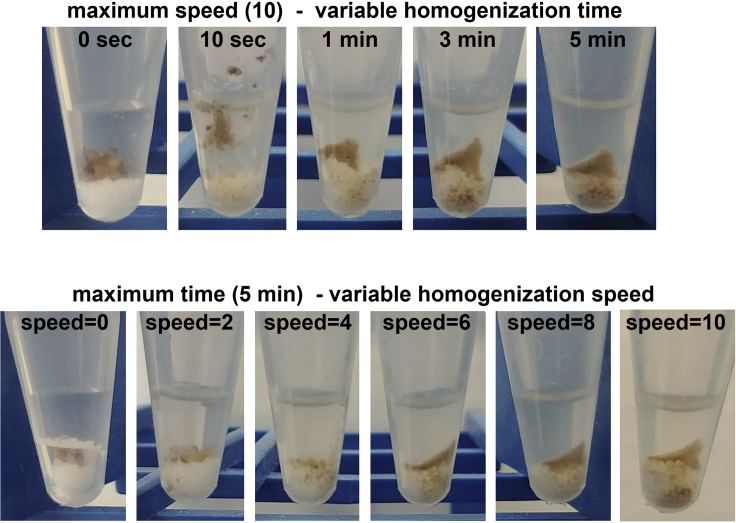
CRITICAL: Homogenization is a crucial step. If the tissue is not homogenized well, tissue fragments (and hence some Triton X-100 soluble proteins) will be pulled into the insoluble fraction, resulting in unreliable experimental results. The use of white-colored beads for the homogenization of dark tissues (like Drosophila thoraces and heads) allows visual determination of the completeness of tissue homogenization. If tissue fragments are still present, the sample should be homogenized again for 1–2 minutes (or more if needed). The sample should be visually inspected to ensure that homogenization is complete (Figure 1). No unneeded homogenization should be done as this increase foaming and the sample temperature and hence the chance for protein degradation.
Figure 1.
Impact of speed and time on homogenization
Representative images of samples each consisting of 5 whole Drosophila homogenized at different speeds and for different time intervals in 150 μL PBS. After homogenization, the samples were briefly centrifuged for 15 s to bring down debris. In the upper panel, the samples are homogenized at maximum speed (10) but for a variable homogenization time (0 s, 10 s, 1 min, 3 min, 5 min). Note the presence of floating, unhomogenized tissues when the homogenization is run for only 10 s. Samples appear to be well homogenized after 3 and 5 min. In the lower panel, the samples are homogenized for a maximum time (5 min) but at variable speeds (0, 2, 4, 6, 8, and 10). Note the presence of unhomogenized tissues (such as red fly eyes) at speed 2 and 4. Samples appear to be well homogenized at speed 6, 8, and 10. Homogenize at the maximum speed (10) and time (5 min) for best results.
-
2.Recover Triton X-100 soluble fractions following centrifugation of homogenate from step 1.
-
a.Homogenates are then centrifuged at 21,000 × g (in a Thermo Scientific Sorvall Legend Micro 21 refrigerated microcentrifuge and rotor #75772446, or equivalent) at 4°C for 10 min to pellet down detergent-insoluble fractions.
-
b.The supernatant is then collected as Triton X-100 soluble fraction.
-
a.
-
3.Wash the pellet with Triton X-100 lysis buffer.
-
a.The remaining pellet, which consists of detergent-insoluble fractions (together with beads), is washed 2–3 times. For each wash, 400 μL to 1 mL Triton X-100 buffer are added to the pellet and the samples centrifuged at 21,000 × g for 5 min at 4°C. The supernatant is then discarded.
-
a.
CRITICAL: Washing is an important step to make sure that the soluble proteins are completely removed before solubilizing the insoluble protein pellet.
-
4.Solubilize the insoluble pellet proteins in 8M urea and 5% SDS buffer to collect Triton X-100 insoluble fraction.
-
a.The pellet is resuspended at 4°C by homogenization for 5 min at highest speed in a tissue homogenizer (0.5-mm zirconium oxide beads are present from the previous step to aid homogenization) with 90 μL RIPA buffer containing 8M urea, 5% SDS, protease inhibitors, and phosphatase inhibitors.
-
b.Subsequently, the sample is centrifuged at 21,000 × g at 4°C for 10 min.
-
c.The supernatant, which consists of the Triton X-100 insoluble fraction, is then collected, and used for subsequent analysis.
-
a.
CRITICAL: Both urea and SDS are toxic chemicals. All personal protective equipment should be used while making solutions.
Note: 20% SDS solution can be made in water and later adjusted to final concentration as per the requirement. Urea buffer should be made fresh and kept at 22°C–25°C. A 30°C incubator can be used to aid urea dissolution in water. While collecting the insoluble protein fractions, the samples can be kept at 22°C–25°C to ease collection as urea precipitates at lower temperatures.
Note: Purity of soluble and insoluble protein fractions can be assessed by using antibodies against proteins present exclusively in the soluble fraction like GAPDH (glyceraldehye 3-phosphate dehydrogenase) or in the insoluble fraction like Histone H3.
Note: Since equal quantities of tissues are used as starting point for homogenization, equal volumes of detergent-soluble and insoluble fractions are used for subsequent analyses. The amount of protein is further validated by using antibodies against α-tubulin or β-actin.
Preparation of detergent-soluble and detergent-insoluble fractions from human brain organoids
Timing: 1–2 h (depending on the number of samples)
Detergent-soluble and insoluble protein fractions from human brain organoids (obtained as described in (Rai et al., 2021) were prepared as described above for Drosophila tissues with a few notable changes. The protocol is described below.
-
5.Homogenize brain organoids in Triton X-100 cell lysis buffer with the aid of a tissue homogenizer.
-
a.Human brain organoids are briefly washed with PBS at 22°C–25°C to remove culture medium, removed from the cultured wells by using pipette tips with trimmed tips, and gently pelleted by centrifugation at 2,000 × g for 5 min. Because of their dimensions, organoids are visible by naked eye and consequently a similar number of organoids (e.g., 2–3) can be pooled into each eppendorf tube.
-
b.50 μL of Triton X-100 lysis buffer is used for homogenization of pools of 2–3 organoids per sample. Homogenization is done for 30 s at maximum speed by using a tissue homogenizer (NextAdvance) and zirconium 0.5-mm beads at 4°C.
-
a.
CRITICAL: Do not homogenize for more than 30 seconds as excessive homogenization can lead to protein denaturation and poor yields of protein fractions.
-
6.Recover Triton X-100 soluble fractions following centrifugation of homogenate from step 5.
-
a.Homogenates from step 5 are centrifuged at 21,000 × g at 4°C for 10 min to pellet down detergent-insoluble fractions.
-
b.The supernatant is then collected as Triton X-100 soluble fraction.
-
a.
-
7.Wash the pellet with Triton X-100 lysis buffer.
-
a.The remaining pellet from step 6 is washed with 400 μL to 1 mL of Triton X-100 buffer and the samples centrifuged at 21,000 × g for 5 min at 4°C. The supernatant is then discarded. The washing is repeated 1–2 times.
-
a.
-
8.Recover Triton X-100 insoluble proteins by solubilizing the pellet in 8M urea and 5% SDS buffer.
-
a.The pellet (along with the beads) from step 7 is homogenized in 50 μL RIPA buffer containing 8M urea, 5% SDS, protease inhibitors, and phosphatase inhibitors.
-
b.Subsequently, the sample is centrifuged at 21,000 × g at 4°C for 10 min.
-
c.The supernatant, which consists of the Triton X-100 insoluble fraction, is then collected, and used for subsequent analysis.
-
a.
Note: Protein concentration in Triton X-100 soluble fractions is quantitated using the BCA assay with BSA (bovine serum albumin) as protein standard. The amount of detergent-soluble protein loaded on SDS-PAGE gels is determined based on the amount of protein quantified in detergent-soluble fractions. However, protein quantitation is not possible from insoluble fractions as the BCA assay reagents are not compatible with urea/SDS. On this basis, the volume of detergent-insoluble protein loaded on SDS-PAGE gels is determined based on the amount of protein quantified in the corresponding detergent-soluble fractions. The amount of protein loaded is further normalized by using antibodies against α-tubulin or β-actin.
Note: Typically, ~15–50 μg of protein can be obtained from 3–4 organoids. If the homogenization is done in 50 μL, this results in ~0.3–1 μg/μL. The sizes of organoids differ and can significantly change the amount of protein retrieved, which makes it imperative to quantify the protein concentration.
Preparation of detergent-soluble and detergent-insoluble fractions from HEK293 cells
Timing: 1–2 h (depending on the number of samples)
Detergent-soluble and insoluble fractions from human HEK293 cells are obtained following the procedures described before for mammalian cells (Holden and Horton, 2009), i.e., with some changes compared to the main protocol described above for Drosophila tissues.
-
9.Resuspend HEK 293 cells in Triton X-100 cell lysis buffer.
-
a.Remove excess media from cell culture wells and leave only 500 μL–1 mL of media. Scrape cells by using a cell scraper (no additional trypsin required) and collect in a 1.5 mL eppendorf tube. Centrifuge at 2,000 × g for 5 min to pellet the cells. Discard any media present in the tube carefully without disturbing the cell pellet.
-
b.HEK293 cell lysis is not done with a tissue homogenizer but rather by resuspending the cells by gentle pipetting in ice-cold Triton X-100 buffer (1% Triton X-100 in PBS containing protease inhibitors and phosphatase inhibitors).
-
a.
-
10.Collect Triton X-100 soluble fractions following centrifugation of homogenate from step 9.
-
a.Homogenates from step 9 are centrifuged at 21,000 × g at 4°C for 10 min to pellet down detergent-insoluble fractions.
-
b.The supernatant is collected as Triton X-100 soluble fraction.
-
a.
-
11.Wash the pellet with Triton X-100 lysis buffer.
-
a.The pellet from step 10 is washed with 400 μL to 1 mL of Triton X-100 buffer and the samples centrifuged at 21,000 × g for 5 min at 4°C. The supernatant is then discarded. The washing is repeated 1–2 times.
-
a.
-
12.Recover Triton X-100 insoluble proteins by solubilizing the pellet in 7M urea and 1% SDS buffer.
-
a.The pellet from step 11 is resuspended in RIPA buffer with 7M Urea and 100 U/mL Benzonase and incubated at 22°C–25°C for 15–20 min to allow digestion of genomic DNA.
-
b.After which, SDS is added to a final concentration of 1%. The detergent-insoluble fraction is further extracted by resuspending the pellet using a tissue homogenizer (without beads) at maximum speed for 30 s. The samples are then centrifuged at 21,000 × g at 4°C for 10 min.
-
c.The supernatant, which consists of the Triton X-100 insoluble fraction, is then collected, and used for subsequent analysis.
-
a.
CRITICAL: Benzonase concentration depends on the density of cells used. If there is a bigger pellet corresponding to more cells, a higher concentration of Benzonase should be used to ensure sufficient DNA degradation. As a reference, the above protocol is based on experiments with ~200,000 cells/well (of a 6-well plate) and cultured for ~4 days before harvest.
CRITICAL: Do not add SDS before or during Benzonase treatment as SDS can denature the enzyme. Add SDS only 15–20 minutes after Benzonase treatment.
Note concerning estimation of protein concentrations: As described in the protocols above, protein concentration is quantitated for the detergent-soluble protein fractions and equivalent volumes of the corresponding detergent-insoluble fractions are then analyzed by SDS-PAGE. The amount of protein loaded is further normalized by using antibodies against α-tubulin or β-actin. A typical protein concentration for detergent-soluble fractions is 1–2 μg/μL (100–200 μg in 100 μL) for ~200,000 cells/well (of a 6-well plate) cultured for ~4 days before harvest.
Preparation of detergent-soluble and detergent-insoluble protein fractions from mouse skeletal muscle
Timing: 1–2 h (depending on the number of samples)
-
13.Homogenize dissected mouse skeletal muscles in NP-40 lysis buffer using a tissue homogenizer
-
a.Mouse muscles are dissected and snap frozen in liquid nitrogen.
-
b.Subsequently, muscle tissues are homogenized in ice-cold NP40 cell lysis buffer (Invitrogen #FNN0021; 200 μL per ~5 mg of mouse muscle tissue), containing protease inhibitors and phosphatase inhibitors, by using a tissue homogenizer (NextAdvance) with zirconium 0.5-mm beads at 4°C at maximum speed for 5 min.
-
a.
-
14.Recover NP-40 soluble fractions following centrifugation of homogenate from step 13.
-
a.The tissue homogenates are subsequently centrifuged at 12,000 × g for 10 min at 4oC to pellet insoluble material.
-
b.Subsequently, the supernatant (NP-40 detergent-soluble protein fraction) is collected.
-
a.
-
15.Wash the pellet with NP-40 buffer
-
a.The remaining pellet from step 14, which consists of NP-40 detergent-insoluble fractions, is washed three times. For each wash, 400 μL to 1 mL ice-cold NP40 cell lysis buffer is added to the pellet and the samples centrifuged at 21,000 × g for 5 min at 4°C. The supernatant is discarded.
-
a.
-
16.Recover NP-40 detergent-insoluble fractions by resuspending the pellet in 8M Urea and 1% SDS buffer.
-
a.8M urea and 1% SDS in PBS (insoluble buffer) is added to solubilize the detergent-insoluble pellet.
-
b.The specific volume added is determined based on the concentration of the corresponding detergent-soluble fraction. For example, if the detergent-soluble fractions of sample A and sample B are 1 mg/mL and 2 mg/mL respectively, then 100 μL of insoluble buffer is added to sample A and 200 μL to sample B.
-
a.
Note: 8M urea dissolved in RIPA buffer can be used in step 16 rather than 8M urea and 1% SDS in PBS.
Note: The protein concentration of detergent-soluble fractions is determined by using standard assays, such as the Bio-Rad protein assay with BSA as a standard (1 mg/mL at highest concentration and ½ serial dilutions); samples are usually diluted 1/10 in water to ensure they are in the range of the BSA standard. The sample concentrations are interpolated into the linear curve for the BSA standard, and the final concentration is determined by factoring in the 1/10 dilution. Detergent-soluble protein fractions are then diluted to matching concentrations using NP40 buffer as diluent. For example, if sample A was 1 mg/mL protein in 100 μL and sample B was 2 mg/mL in 100 μL then 100 μL of NP40 buffer should be added to sample B, such that both samples now have a matching concentration of 1 mg/mL.
Note: Utilization of NP40 cell lysis buffer is more amenable to certain follow-up applications such as immunoprecipitation.
Note: Certain tissues such as heart, spleen, and bone, are more difficult to homogenize. While the procedure described above works optimally for skeletal muscle, a modification of the protocol could include the utilization of a different set of beads, such as the NAVY bead lysis kit (NextAdvance), which consists of microcentrifuge tubes pre-filled with a mix of stainless-steel beads of different sizes.
Note: Typically, ~1 mg of protein can be obtained from the detergent-soluble fraction from 10 mg of muscle tissue (mouse tibialis anterior muscle).
SDS-PAGE of detergent-soluble and detergent-insoluble fractions
Timing: 2–3 h of work (depending on the number of blots) distributed over several days.
-
17.
Sample preparation for SDS-PAGE
Protein samples are combined with SDS blue loading buffer containing DTT (Cell signaling #7722, as per user’s manual) and boiled at 95°C for 5 min.
-
18.
The samples are centrifuged briefly to collect the sample to the bottom of the tube and then loaded into 4–20% gradient polyacrylamide gels (Bio-Rad) with a protein weight marker in one well.
-
19.
Subsequently, standard procedures for SDS-PAGE and western blotting are used. Typically, the gels run for 1.5–2 h at 100 V and transfer is done at 100 V for 1 h.
-
20.
Blotting conditions are done following standard protocols, including typical dilutions of 1:1000 for primary antibodies and 1:5000 for secondary antibodies.
Note: Typically, 5 μg of proteins for detergent-soluble fractions and an equal volume of detergent-insoluble fraction (corresponding to the detergent-soluble fraction) are examined by SDS-PAGE.
Data collection and analysis
Timing: <1 h of work (depending on the number of blots)
Once the western blots are developed, the intensity of the bands can be estimated with a number of software programs, including the Histogram function of Photoshop, NIH ImageJ, and others. Multiple independent experiments with the same or related experimental interventions and/or side comparison of biological replicates is recommended.
Statistical analysis is done with appropriate standard tests. For example, the unpaired two-tailed Student’s t-test is used to compare the means of two independent groups to each other whereas one-way ANOVA with Tukey’s post hoc test is used for multiple comparisons of more than two groups of normally distributed data.
Expected outcomes
The western blot analysis of detergent-soluble and detergent-insoluble fractions allows for the identification of interventions (genetic, pharmaceutical, dietary, etc.) that modify the age-related accumulation of poly-ubiquitinated proteins and of other aggregation-prone proteins. Moreover, this process separates detergent-insoluble proteins (which correspond to aggregates of misfolded proteins) and detergent-soluble proteins (which correspond to soluble proteins undergoing normal turnover and misfolded proteins en route to degradation, primarily by the ubiquitin proteasome system), which allows for further analysis of each class of protein individually using other markers.
It is expected that aging and disease conditions that are characterized by reduced protein degradation (i.e., that have dysfunctional ubiquitin-proteasome and autophagy-lysosome systems) will result in accumulation of poly-ubiquitinated proteins and other misfolded and/or aggregation-prone proteins, which are then typically sequestered by the cells into dedicated subcellular compartments, known as aggresomes or protein aggregates (DiLoreto and Murphy, 2015). This increase in the amount of poly-ubiquitinated proteins can also be detected in the detergent-soluble fractions: these may consist of poly-ubiquitinated proteins en route to aggresomes or en route to degradation by the ubiquitin-proteasome system.
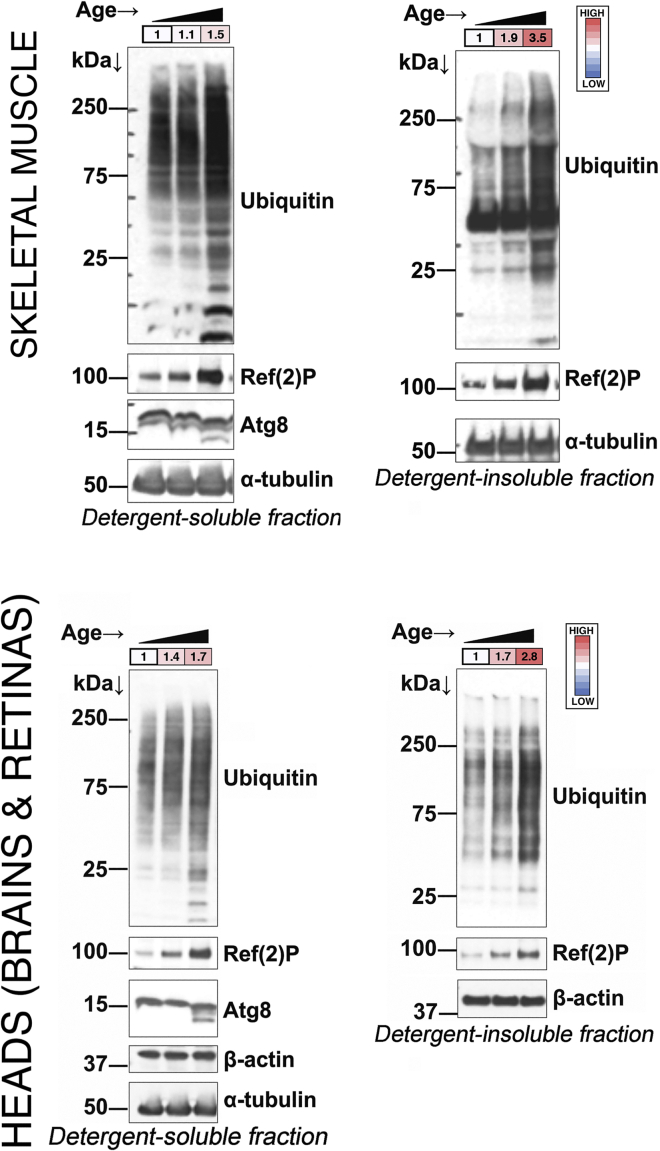
Figure 2 reports an example of western blot analyses of detergent-soluble and detergent-insoluble protein fractions obtained from heads (brains and retinas) and thoraces (which consist mostly of muscle) of Drosophila at different ages. As expected, there is an age-associated increase in the amount of poly-ubiquitinated proteins found in detergent-insoluble fractions, consistent with the notion that there is pervasive decline in proteostasis across tissues during aging in Drosophila (Rai et al., 2021).
Figure 2.
Age-related increase in detergent-soluble and -insoluble polyubiquitinated proteins in Drosophila
Aging leads to an increase in poly-ubiquitinated proteins found in detergent-insoluble (and to a lower extent in detergent-soluble) fractions of Drosophila tissues, such as the fly thorax (which consists primarily of skeletal muscle) and the head (which primarily consists of retina and brain). Age-related increase in poly-ubiquitinated proteins is accompanied by parallel increases in the levels of Ref(2)P, the Drosophila homolog of p62/SQSTM1. Moreover, the ratio of LC3/Atg8-II (lower band) versus LC3/Atg8-I declines with aging, which is presumably indicative of an age-related decline in the autophagic flux; β-actin and/or α-tubulin are shown as normalization controls. The quantitation of ubiquitin (full lane) to the α-tubulin or β-actin band is shown in colored boxes. This data is taken from reference (Rai et al., 2021).
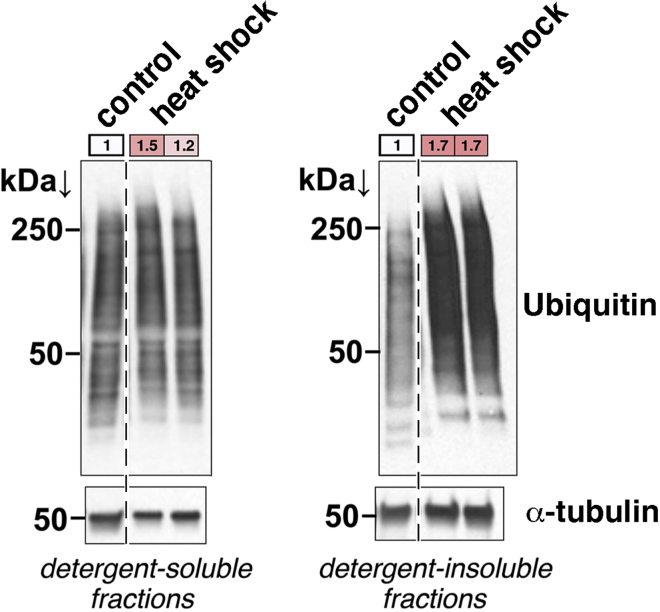
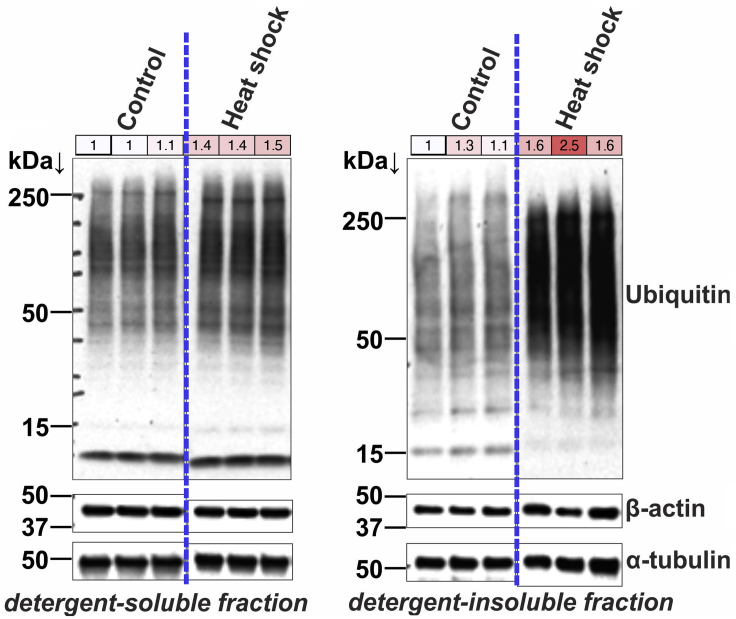
In addition to aging and to diseases characterized by proteostatic decline, this technique can also be used to estimate the impact of cellular stress in human brain organoids (Figure 3) and human HEK293 cells (Figure 4). Heat shock of brain organoids and HEK293 cells results in an increase in the poly-ubiquitinated proteins found in the insoluble fraction, indicating an accumulation of protein aggregates upon heat shock (Rai et al., 2021).
Figure 3.
Increase in detergent-soluble and -insoluble polyubiquitinated proteins in human brain organoids upon heat shock
Heat shock (41.5°C for 7 h) leads to an increase in poly-ubiquitinated proteins found in detergent-insoluble (and to a lower extent in detergent-soluble) fractions from cultured human brain (cortical) organoids. The quantitation of ubiquitin (full lane) to the α-tubulin band is shown in colored boxes. The dashed line separates lanes taken from the same western blot membrane. This data is taken from reference (Rai et al., 2021).
Figure 4.
Increase in detergent-soluble and -insoluble polyubiquitinated proteins in HEK293 cells upon heat shock
HEK293 cells were subjected to heat shock at 41.5°C for 7 h. As with cortical organoids, heat shock leads to an increase in poly-ubiquitinated proteins in TritonX 100-insoluble fractions. The quantitation of ubiquitin to α -tubulin is shown in colored boxes. The dashed line separates lanes taken from the same western blot membrane. This data is taken from reference (Rai et al., 2021)
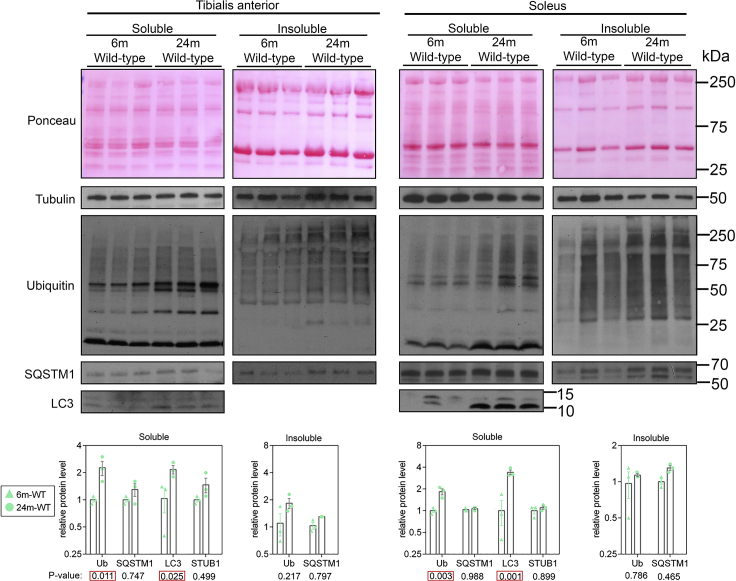
Apart from invertebrate and cell/organoid models, the methods here described can be used to determine the status of proteostasis in tissues from various disease models and organisms, such as mice. In Figure 5, we present an example of changes in proteostasis that occur with aging in mouse skeletal muscles. Specifically, with aging there is an increase in poly-ubiquitinated proteins found in the insoluble fractions of skeletal muscles from 24-month-old mice as compared to muscle from young 6-month-old mice (Hunt et al., 2021), consistent with other studies (Sakuma et al., 2016; White et al., 2016).
Figure 5.
Decline in protein quality control in skeletal muscles during aging
Western blotting of detergent-soluble and insoluble proteins fractions from tibialis anterior and soleus muscles from wild-type mice at 6 months (young) and 24 months (old) of age. Note the overall age-related increase in poly-ubiquitinated proteins found in detergent-soluble and -insoluble fractions. Age-related increase in poly-ubiquitinated proteins is accompanied by parallel increases in the levels of p62/SQSTM1 and in the ratio of LC3-II (lower band) versus LC3-I, which is presumably indicative of decline in the autophagic flux with aging. Ponceau and α-tubulin are shown are normalization controls. The quantitation (mean values ±SEM) and statistical analysis (p-values) are shown. This data is taken from reference (Hunt et al., 2021).
Limitations
As for most biological techniques, the western blot analysis of detergent-soluble and insoluble fractions provides valuable information that should be corroborated with complementary experimental approaches. For example, immunostaining with the same tools (antibodies for p62 and poly-ubiquitinated proteins) and confocal microscopy can be used to identify age-related cellular/tissue changes (Hunt and Demontis, 2013; Hunt et al., 2021; Rai et al., 2021).
The analysis with western blot, qRT-PCR, and immunostaining can also indicate whether proteostasis regulators, such as the autophagy proteins Ref(2)P/p62 and Atg8/LC3, are modified with aging or in the specific disease context examined.
Troubleshooting
Problem 1
Insufficient recovery of protein in the detergent-soluble and/or detergent-insoluble fractions (steps 1, 4, 5, 8, 9, 13, 16).
Potential solution
Ensure that tissue homogenization is optimally done by visually inspecting the sample after homogenization (Figure 1). If the homogenization is optimal but the issue persists, increase the amount of tissue used if required.
Tip: Homogenize the tissues and observe with naked eyes whether any chunks of tissues or whole tissues are present. Homogenize again until no visible tissue chunks are left. If in doubt, homogenize at the maximum speed and time.
Tip: The efficiency/quality of the homogenization can be further tested by probing for protein markers such as GAPDH and histone H3 that are enriched in the detergent-soluble and detergent-insoluble fractions, respectively.
Problem 2
Insufficient recovery of protein in the detergent-insoluble fractions .
Potential solution
If there is optimal recovery of detergent-soluble proteins but not of detergent-insoluble proteins, ensure that the stress conditions used are sufficient to perturb proteostasis as limited recovery of detergent-insoluble proteins may be a result of insufficient thermal stress (for cell culture or organoid experiments) or animals used were not old enough.
Tip: 7 h of heat shock at 41.5°C for cells or organoids is sufficient to perturb proteostasis. For Drosophila reared at 25°C, 10 days is considered young whereas 30 and 50–60 days are considered middle and old age, respectively. As a rough estimate, mice are considered young at up to 6 months of age and considered old at 20–24 months or older ages. Proteostasis is compromised in old animals compared to young or middle-aged organisms.
Problem 3
Insufficient recovery of proteins in the detergent-insoluble fractions of HEK293 cells or sticky insoluble fractions (step 12).
Potential solution
This could occur because of inadequate DNA enzymatic digestion. Repeat the extraction with a higher concentration of Benzonase (1.5–2 times higher).
Problem 4
Inconsistent protein ubiquitination pattern (steps 1–16).
Potential solution
If the soluble fraction is not extracted efficiently due to poor homogenization or the insoluble protein pellet is not washed enough to get rid of soluble proteins completely, there is a possibility that soluble proteins are reflected in the insoluble fractions. Repeat the experiment by making sure the tissues are well homogenized prior to extraction of soluble proteins and that the pellet is washed properly.
Problem 5
High background on the western blot membrane post antibody probing (step 20).
Potential solution
This could be a result of high concentrations of primary antibody. Optimize antibody concentrations followed by stringent washes. Another reason could be poor membrane blocking. Anti-ubiquitin (Santa Cruz#8017) works best with 5% BSA as blocking agent.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Fabio Demontis (Fabio.Demontis@stjude.org).
Materials availability
This study did not generate unique reagents.
Data and code availability
This study did not generate new data and codes.
Acknowledgments
This work was supported by research grants to F.D. from the Ellison Medical Foundation, Glenn Foundation for Medical Research, American Federation for Aging Research, Hartwell Foundation, American Parkinson Disease Association, and the National Institute on Aging of the NIH (R01AG055532 and R56AG063806). L.C.H. was supported by a Glenn/AFAR Postdoctoral Fellowship for Translational Research on Aging. Research at St. Jude Children’s Research Hospital is supported by the ALSAC. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
M.R., L.C.H., and F.D. wrote the manuscript with input from M.C., Z.C., A.N., J.J., and F.A.G.
Declaration of interests
The authors declare no competing interests.
References
- Demontis F., Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F., Piccirillo R., Goldberg A.L., Perrimon N. Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis. Model. Mech. 2013;6:1339–1352. doi: 10.1242/dmm.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLoreto R., Murphy C.T. The cell biology of aging. Mol. Biol. Cell. 2015;26:4524–4531. doi: 10.1091/mbc.E14-06-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P.M., Dillin A. Protein homeostasis and aging in neurodegeneration. J. Cell Biol. 2010;190:719–729. doi: 10.1083/jcb.201005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden P., Horton W.A. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res. Notes. 2009;2:243. doi: 10.1186/1756-0500-2-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L.C., Demontis F. Whole-mount immunostaining of Drosophila skeletal muscle. Nat. Protoc. 2013;8:2496–2501. doi: 10.1038/nprot.2013.156. [DOI] [PubMed] [Google Scholar]
- Hunt L.C., Jiao J., Wang Y.D., Finkelstein D., Rao D., Curley M., Robles-Murguia M., Shirinifard A., Pagala V.R., Peng J. Circadian gene variants and the skeletal muscle circadian clock contribute to the evolutionary divergence in longevity across Drosophila populations. Genome Res. 2019;29:1262–1276. doi: 10.1101/gr.246884.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L.C., Schadeberg B., Stover J., Haugen B., Pagala V., Wang Y.D., Puglise J., Barton E.R., Peng J., Demontis F. Antagonistic control of myofiber size and muscle protein quality control by the ubiquitin ligase UBR4 during aging. Nat. Commun. 2021;12:1418. doi: 10.1038/s41467-021-21738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L.C., Stover J., Haugen B., Shaw T.I., Li Y., Pagala V.R., Finkelstein D., Barton E.R., Fan Y., Labelle M. A Key Role for the Ubiquitin Ligase UBR4 in Myofiber Hypertrophy in Drosophila and Mice. Cell Rep. 2019;28:1268–1281.e6. doi: 10.1016/j.celrep.2019.06.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L.C., Xu B., Finkelstein D., Fan Y., Carroll P.A., Cheng P.F., Eisenman R.N., Demontis F. The glucose-sensing transcription factor MLX promotes myogenesis via myokine signaling. Genes Dev. 2015;29:2475–2489. doi: 10.1101/gad.267419.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Demontis F. Skeletal muscle autophagy and its role in sarcopenia and organismal aging. Curr. Opin. Pharmacol. 2017;34:1–6. doi: 10.1016/j.coph.2017.03.009. [DOI] [PubMed] [Google Scholar]
- Labbadia J., Morimoto R.I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M., Coleman Z., Curley M., Nityanandam A., Platt A., Robles-Murguia M., Jiao J., Finkelstein D., Wang Y.D., Xu B. Proteasome stress in skeletal muscle mounts a long-range protective response that delays retinal and brain aging. Cell Metab. 2021;33:1137–1154. doi: 10.1016/j.cmet.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma K., Kinoshita M., Ito Y., Aizawa M., Aoi W., Yamaguchi A. p62/SQSTM1 but not LC3 is accumulated in sarcopenic muscle of mice. J. Cachexia Sarcopenia Muscle. 2016;7:204–212. doi: 10.1002/jcsm.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White Z., White R.B., McMahon C., Grounds M.D., Shavlakadze T. High mTORC1 signaling is maintained, while protein degradation pathways are perturbed in old murine skeletal muscles in the fasted state. Int. J. Biochem. Cell Biol. 2016;78:10–21. doi: 10.1016/j.biocel.2016.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new data and codes.