Fig. 5.
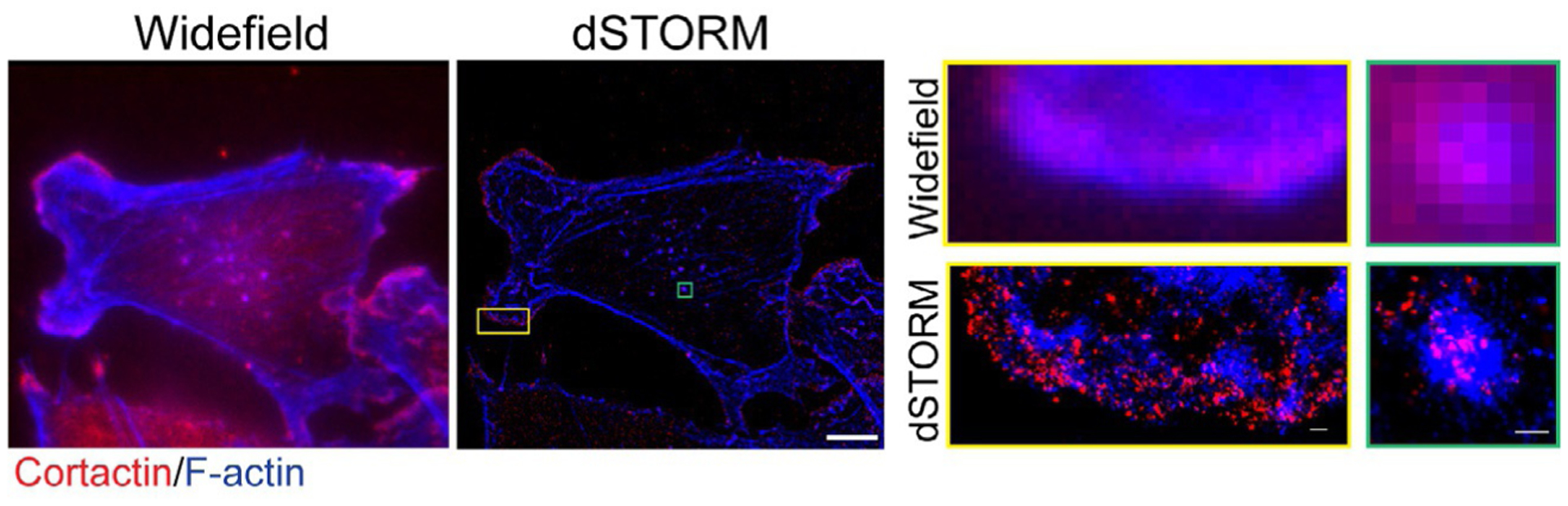
dSTORM image of a breast cancer cell. (Left panel): Widefield image of a cancer cell on gelatin matrix. F-actin in blue, cortactin in red. (Middle panel): dSTORM image of the same cancer cell. Top and bottom inset #1 (yellow box): Widefield and dSTORM of a lamellipodia structure. Top and bottom inset #2 (green box): Widefield and dSTORM showing spatial distribution of F-actin and cortactin in an invadopodia structure. Left and middle panel, scale bar: 10μm. Insets, scale bar: 1μm. Data acquisition for dSTORM was carried out on the Nanoimager S (Oxford NanoImaging, ONI, Oxford, UK). Signals from Alexa 647 and Alexa 488 were recorded sequentially for 10,000 frames each. Localization and image rendering were performed in the NimOS v1.4 software, and the final reconstruction displayed in a precision mode.
