FIGURE 5.
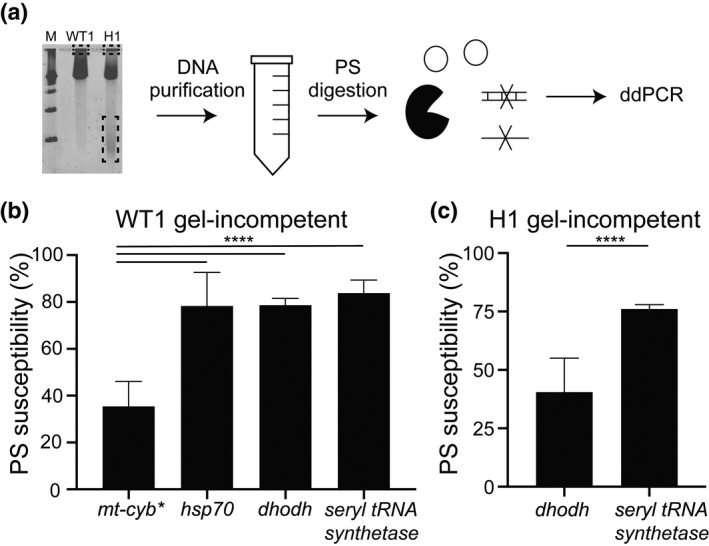
Exonuclease digestion indicates structural conservation between the complex mitochondrial genome and extrachromosomal DNA. (a) Following PFGE (as in Figure 2b), specific regions of the agarose gel were excised (as in Figure 4), DNA was purified, pre‐treated with a frequent cutting restriction enzyme (not depicted), digested with the PS exonuclease, and assessed by Droplet Digital (dd)PCR for specific genomic loci (either single or multi‐copy, Table 1). For simplicity, circular molecules represent both complex and circular DNA elements. (b and c) The PS susceptibility of each genomic locus was determined by quantifying DNA‐containing droplets using ddPCR; the % PS susceptibility (or level of DNA degradation) was calculated by dividing the number of loci‐positive droplets after digestion by the number of loci‐positive droplets before digestion and multiplying by 100. Single copy genes:heat shock protein(hsp70) andseryl‐tRNA synthetase; multi‐copy genes: mitochondrialcytochrome b(mt‐cyb*)anddihydroorotate dehydrogenase(dhodh, Table 1). *, although the majority ofP. falciparummitochondrial genomes are linear, complex structural forms are entrapped in the PFGE loading well. Error bars denote standard error; ****p < 0.0001. (b) DNA derived from the loading well of WT1 (Dd2, gel‐incompetent DNA, N = 3). (c) DNA derived from the loading well of the high‐level resistant clone H1 (gel‐incompetent DNA, N = 3)
