Fig 1.
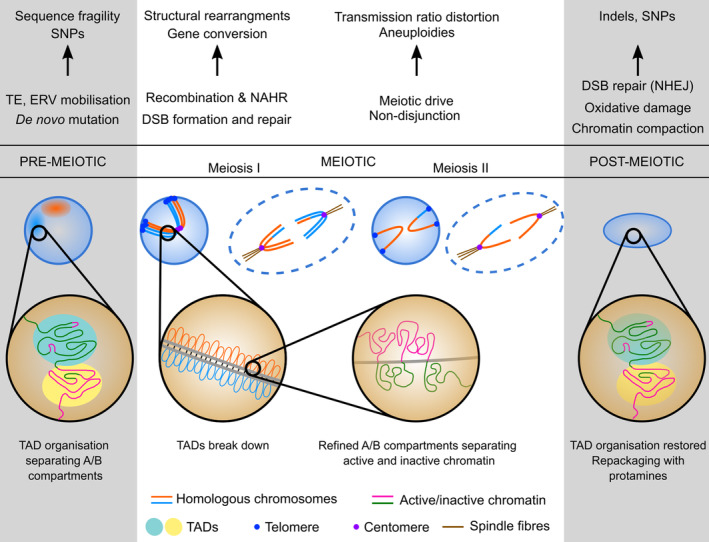
Illustration of genomic reconfiguration during spermatogenesis, and the processes that can lead to mutational outcomes. Prior to meiosis, chromatin is organised into topologically associated domains (TADs). These TADs are lost during prophase I, wherein the chromatin is separated more distinctly into active and inactive compartments despite its aggregation into a linear array of loops bound along a proteinaceous chromosome axis. Mobilisation of transposable elements (TEs) and endogenous retroviruses (ERVs), along with programmed recombination depending on the topoisomerase‐related enzyme, meiotic recombination protein SPO11 and nuclear division, provide opportunities for structural genomic changes via non‐allelic homologous recombination (NAHR) or non‐dysjunction, while the repair of DNA double‐strand breaks (DSBs) at the post‐meiotic stage also allows smaller indels and single nucleotide polymorphisms (SNPs) to arise via the non‐homologous end joining (NHEJ) pathway. Data from Alavattam et al. (2019), Patel et al. (2019) and Wang et al. (2019).
