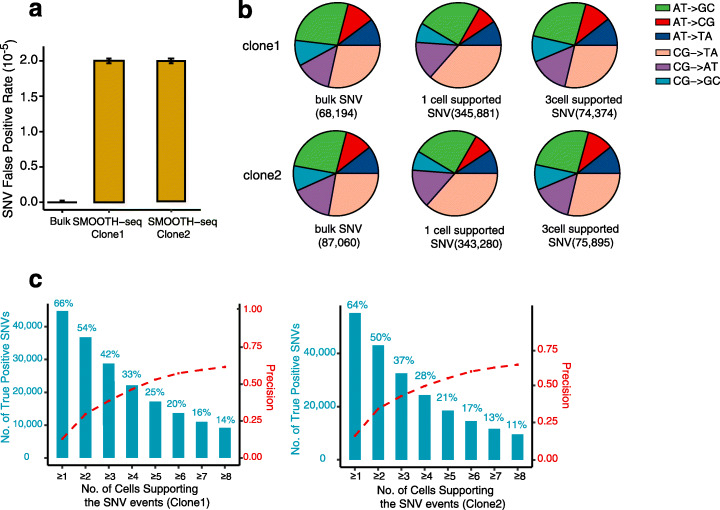
Fig. 4.
SNV detection in K562 cells. a False positive rate (FPR) of SMOOTH-seq in detecting SNVs in single K562 cells. The error bar generates according to the values in 95 single cells. The bulk SNVs are regarded as the standard reference with the FPR as 0. b Spectra of SNVs in bulk and SMOOTH-seq of K562 genome. SMOOTH-seq exhibit slightly biased CG-to-TA false positives in one K562 cells. c Precision of SMOOTH-seq detecting SNVs and the percentage of true positive SNVs with different numbers of supporting clone 1 or clone 2 K562 cells