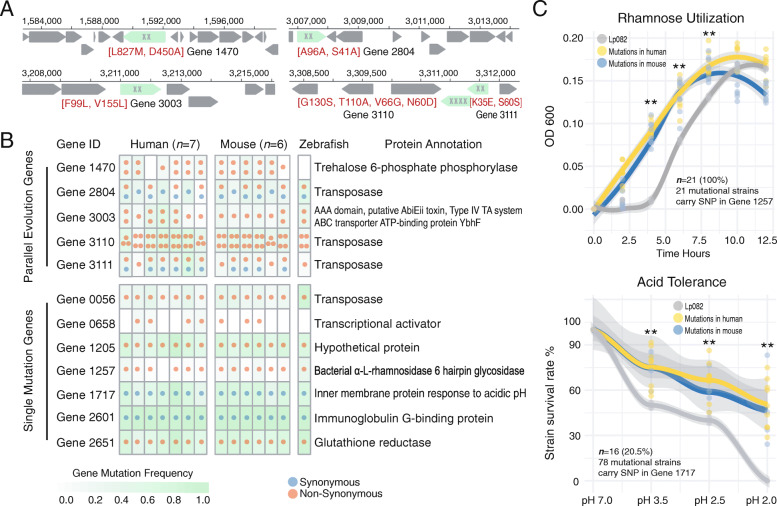
Fig. 2.
Genes that underwent in vivo evolution and the enhanced competitive fitness of the probiotic in mammalian hosts. A The locations of the five parallel evolutionary genes in the Lp082 chromosome. The genes highlighted by green color represent the parallel evolutionary genes and the “x” symbols represent the number of mutations detected in the gene. Annotations in red color represent the mutations that lead to functional changes in the amino acid sequence (i.e., L(Leu) to M(Met)). B Five probiotic genes underwent parallel evolution in different host species, while the other seven genes only have a single mutation. Each dot in the grid chart represents an independent mutation event and is also colored by types of mutation (synonymous or non-synonymous). The color within each cell represents the mutation frequency of a gene in all probiotic isolates from a host subject. C Either non-synonymous or synonymous SNPs can confer the growth benefits of probiotics in the host gut. The phenotypic verification experiments of probiotic isolates related to the rhamnose utilization (top panel) and acid tolerance (bottom panel). A non-synonymous SNP identified in Gene 1257 (annotated as bacterial alpha-L-rhamnosidase 6 hairpin glycosidase) from 21 mutational isolates (100%). This SNP conferred growth benefits to all isolates by utilizing the rhamnose more rapidly than the original strain. Remarkably, a synonymous SNP in Gene 1717 (annotated as inner membrane protein response to acidic) was found in 78 isolates. Among these, 16 mutational isolates (20.5%) improved the performance of this strain in acid tolerance regardless of a synonymous SNP