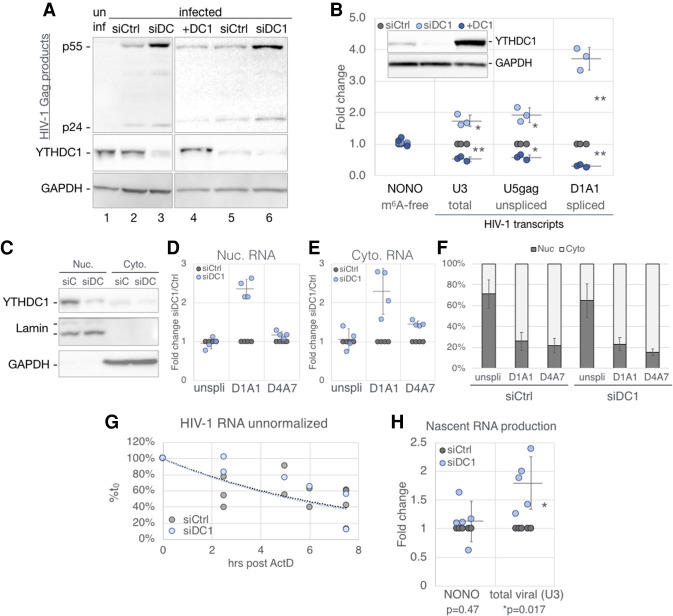
Figure 5.
YTHDC1 regulates HIV-1 gene expression with no detectable effect on RNA nuclear export. 293T cells transfected with nontargeting (siCtrl, gray) or YTHDC1-targeting (siDC1, light blue) siRNAs or were transfected with a YTHDC1 expression vector (+DC1, dark blue) or empty vector. HIV-1 single-cycle infections were performed for the following analyses: (A) Viral Gag protein expression assayed by Western blot, costained with a YTHDC1 antibody. (B) Viral RNA levels assayed by qRT-PCR, n = 3, with the m6A-free host NONO mRNA as a control. A representive Western blot shown in the top left inset depicts the validation of YTHDC1 levels for samples used in this panel. (C–F) Subcellular fractionation assay of infected siCtrl and siDC1 cells. (C) Western blot validation of fractionation, stained for YTHDC1, nuclear Lamin A/C, and cytosolic GAPDH. (D–F) HIV-1 transcript alternatively spliced isoforms were quantified by qRT-PCR with primers targeting unspliced (unspli, U5-gag), and the D1/A1 and D4/A7 splice junctions, calculated as fold change of siDC1 over siCtrl in the nuclear (D), and cytosolic fraction (E). (F) The same RNA quantification as in D and E calculated as percent nuclear and percent cytoplasmic. (G) Stability of RNA transcripts in siCtrl and siDC1 cells assayed by treating infected cells at 2 dpi with ActD and the viral RNA levels from the indicated time points analyzed by qRT-PCR and shown as percentage of the RNA level at time point 0. (H) Production of nascent HIV-1 transcripts in infected siCtrl and siDC1 cells was measured by pulsing cells with the nucleoside analog 4SU for 1.5 h. The 4SU+ transcripts were then isolated and quantified by qRT-PCR. Statistical analysis used Student's t-test. Error bars indicate SD. (*) P < 0.05, (**) P < 0.01.