Abstract
Introduction
The SARS-CoV-2 pandemic has led to one of the most critical and boundless waves of publications in the history of modern science. The necessity to find and pursue relevant information and quantify its quality is broadly acknowledged. Modern information retrieval techniques combined with artificial intelligence (AI) appear as one of the key strategies for COVID-19 living evidence management. Nevertheless, most AI projects that retrieve COVID-19 literature still require manual tasks.
Methods
In this context, we present a novel, automated search platform, called Risklick AI, which aims to automatically gather COVID-19 scientific evidence and enables scientists, policy makers, and healthcare professionals to find the most relevant information tailored to their question of interest in real time.
Results
Here, we compare the capacity of Risklick AI to find COVID-19-related clinical trials and scientific publications in comparison with clinicaltrials.gov and PubMed in the field of pharmacology and clinical intervention.
Discussion
The results demonstrate that Risklick AI is able to find COVID-19 references more effectively, both in terms of precision and recall, compared to the baseline platforms. Hence, Risklick AI could become a useful alternative assistant to scientists fighting the COVID-19 pandemic.
Keywords: Artificial intelligence, COVID-19, Search platform, Risklick
Introduction
The SARS-CoV-2 pandemic resulted in one of the largest waves of publications and clinical trials in the history of modern science, with the number of articles doubling every 20 days and unprecedented clinical trial rate [1, 2, 3]. In this context, it has become virtually impossible for scientists, policy makers, and healthcare workers to keep up with the speed at which data are generated. Moreover, this situation limited the possibilities offered to professionals involved in the pandemic to read entire articles thoroughly, as well as to properly evaluate the limitations of the data. In addition, this outburst of publications also impacted the average quality of research studies [4, 5].
The necessity to effectively gather scientific evidence that encompasses only relevant information with acceptable quality has been one of the most important modern challenges in science. This issue has become strikingly evident throughout the COVID-19 crisis. In this context, artificial intelligence (AI) appears to be the best strategy to seek the most relevant scientific evidence in a minimum amount of time [6, 7]. AI-based strategies are now required to diminish time of research, increase performance, and reduce errors and oversights in the research of references performed by scientists and health professionals. The proliferation of AI-based initiatives to address the COVID-19 pandemic has resulted in the creation of numerous technologies, such as LitCovid and the COVID-NMA Project, among others [8, 9]. However, most of the developed COVID-19 tools, such as the cited examples, require manual steps in the analytic process. Hence, a fully automated, AI-based efficient tool is still missing in the context of the COVID-19 pandemic in order to optimize the access and the management of specific knowledge and research results.
In this context, we developed a novel, automated, scientific evidence management platform called Risklick AI. The tool aims to gather and manage COVID-19-related literature using natural language processing (NLP), a technology allowing computers to process and analyze large amounts of data expressed in natural language [10, 11, 12]. The tool combines classic statistical word frequency methods, the so-called bag-of-words, with state-of-the-art masked language models [13, 14, 15]. Hence, using AI, Risklick AI allows computers to analyze human language with more meaning than with the usual processed and programed responses. In this study, we compare the capacities of Risklick AI to find COVID-19-related clinical trials compared to clinicaltrials.gov [16] and scientific publications in comparison with PubMed. We compared query outcomes of Risklick AI to clinicaltrials.gov and PubMed on COVID-19 pharmacologically relevant treatments, as considered by the authorities [17]. Here, we demonstrate that Risklick AI represents the more effective technology with the potentiality to assist scientists in finding and pursuing relevant COVID-19-related scientific evidences.
Methods
Data Collection
On a daily basis, Risklick AI collects and updates clinical trial data from wide sources such as clinical trial registries and datasets from the WHO [18]. Moreover, publications' metadata like titles, abstracts, journal names, publication date, digital object identifier number, and others are collected and updated from sources like PubMed, Embase, BioRxiv, and MedRxiv from “Living Evidence on COVID-19” and “CORD-19.”
Technology
All the data for clinical trials and publications are preprocessed to align to a predefined data format and added to Elasticsearch, which serves as a full-text search and analytics engine for clinical trials and publications. The indexed data and queries are normalized using a pipeline of text preprocessing techniques like tokenization, lowercasing, stop words removal, and reducing words to their root form. The indices are maintained in an Elasticsearch cluster. The index model parameters are tuned using a set of manually annotated queries. The similarity measure was computed using the divergence from the randomness model with the term frequency normalization set to 20.0 [19]. A detailed description of the pipeline is provided by Ferdowsi et al. [15].
To increase the recall of relevant documents to the user query, we apply query expansion techniques using a COVID-specific ontology of standardized medical terms, their synonyms, classes, and subclasses engineered by clinical trial domain experts [20]. For instance, once the user searches for heparin, the query automatically expands to all 3 majors of heparin: unfractionated heparin, low-molecular-weight heparin, ultra-low-molecular weight heparin, and their trade names based on COVID-specific ontology (e.g., nadroparin, Fraxiparine, Fraxodi, Calciparine, bemiparin, Zibor, Ivor, enoxaparin, Clexane, Lovenox, Fragmin, Dalteparin, and dociparstat).
Experimental Setup
At the time of analysis, >1,800 interventional studies linked to COVID-19 were available on clinicaltrials.gov. In addition, >48,500 COVID-19-related publications were available on PubMed. In order to compare Risklick AI's performance with other COVID-related search platforms, we defined and used a common set of search queries, which were executed on a specific day for all platforms. To assess our clinical trial search engine, we compared Risklick AI with the most advanced and biggest clinical trial registry − clinicaltrials.gov. In addition to COVID-19 cases, clinicaltrials.gov covers all of COVID-19 clinical trials from other registries like clinicaltrialsregister.eu and chichtr.org, as specified by clinicaltrials.gov (https://clinicaltrials.gov/ct2/who_table). Hence, clinicaltrials.gov appears as the adapted gold standard to allow comparison with Risklick AI.
The day the queries are run, the platform retrieves the latest dataset from clinicltrials.gov, and it is indexed in the Risklick AI platform. The comparison comprises only interventional clinical trials having a unique clinicaltrials.gov identifier (NCT number). To compare the clinical trials found by the different types of queries, data from Risklick AI, clinicaltrials.gov, and corona-trials.org are collected for categories like antibiotic, anticoagulant, and antiviral, as well as more fine granular queries for specific drugs like remdesivir, tocilizumab, azithromycin, hydroxychloroquine, and heparin (see online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000515908).
Risklick AI and PubMed were then compared regarding their publication search performance. Before running the query, the latest COVID-19-related publications are retrieved from PubMed using the predefined queries in the Institute of Social and Preventive Medicine (ISPM), Bern, and added to a new index in Risklick AI [21]. This way we ensure that the queries executed on a specific day on PubMed and Risklick AI retrieve publications based on the same data distribution for the specific day on both platforms. To compare the scientific publications found by the different queries, data from Risklick AI and PubMed were collected for antithrombotic, dexamethasone, and favipiravir (online suppl. Table 2). All the drug categories used in this study (antibiotic, antithrombotic, antiviral, and anticoagulant) are resumed in online suppl. Table 3.
Validation
Verification and validation procedures were performed by 2 separate and independent immunologists. All clinical trials and scientific publications were analyzed and verified manually. To optimize the result comparison between the different search tools, recall (the number of positive class predictions made out of all positive examples in the dataset), precision (the number of positive class predictions that actually belong to the positive class), and F1 score (single score that balances both the concerns of precision and recall in one number) were calculated [22].
Data Analysis
Retrieved publications were individually and manually scored as true-positive or false-positive. Graphs were created using Prism 8.0.
Results
Comparison Search Performance for Clinical Trials
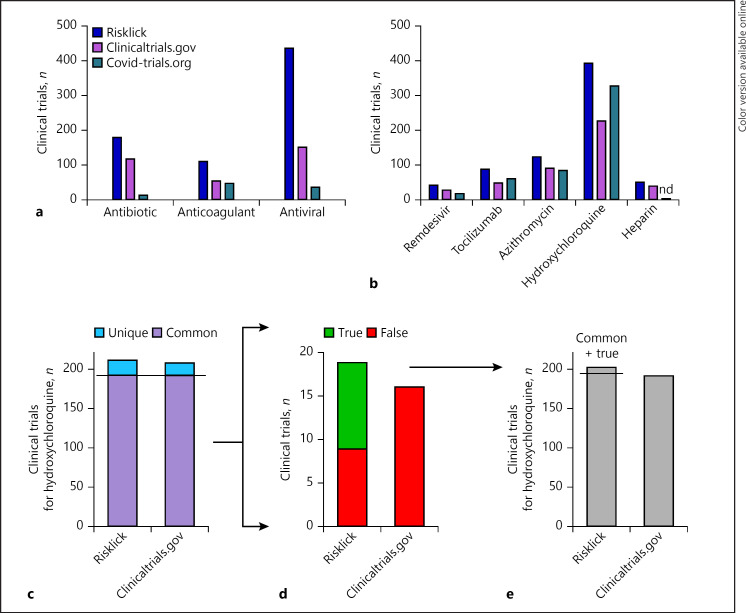
The capacity of Risklick AI to retrieve COVID-19-related clinical trials was analyzed. When compared to clinicaltrials.gov and covid-trials.org, regarding its capacity to find COVID-19-related clinical trials, Risklick AI found more raw clinical trials than other tools for different categories of treatments such as antibiotic, anticoagulant, and antiviral (Fig. 1a). In average, Risklick found 1.9-times more clinical trials than clinicaltrials.gov for these 3 treatments and 8.2-times more than covid-trials.org for these same 3 treatments. When investigating key molecules of each category, such as hydroxychloroquine, remdesivir, azithromycin, tocilizumab, or heparin (Fig. 1b), Risklick AI presented more raw output compared to the 2 other research tools. No clinical trial connected to COVID-19 was found (nd) on covid-trials.org for the heparin query.
Fig. 1.
Risklick AI clinical trial outcome for COVID-19 compared to other web-based resource registries. a, b The total raw number of clinical trials found by Risklick AI compared to other registries for drug classes (a) and specific treatments (b) used against COVID-19. c−e Analysis of search capacity of registered clinical trials by Risklick AI and clinicaltrials.gov based on the same dataset for hydroxychloroquine. Clinical trials were separated between common and unique outcomes (c). Unique outcomes were validated and separated between true-positive (true) and false-positive (false) results (d). The final total number of true-positive clinical trials comprises the addition of common findings and unique, true-positive findings (e). nd, no data.
In order to compare the search capacity of Risklick AI in comparison with clinicaltrials.gov, COVID-19-related search was restricted to the same database, using only clinical trials registered on clinicaltrials.gov. This strategy was applied for both drug classes and specific drugs. Using hydroxychloroquine data for illustration, we first segregated publications found only by Risklick AI or clinicaltrials.gov (“unique”) from publications found by both tools (“common”) (Fig. 1c). Then, unique publications were analyzed and separated between true-positive (“true”) and false-positive (“false”) results (Fig. 1d). Ultimately, we calculated the total number of true-positives of the publications by adding the categories common and unique along with true-positive (Fig. 1e).
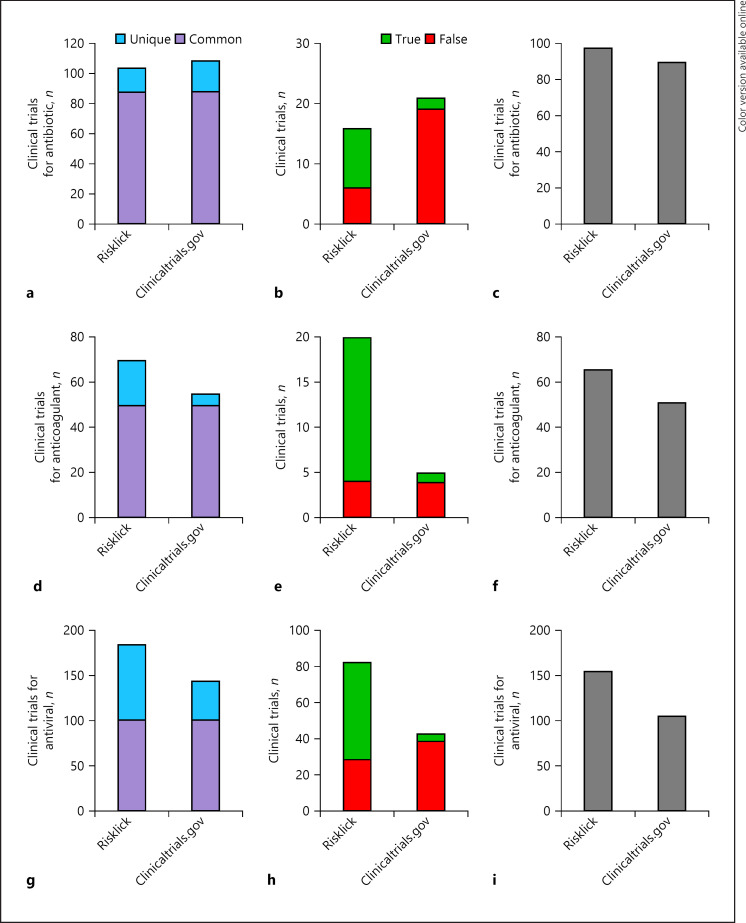
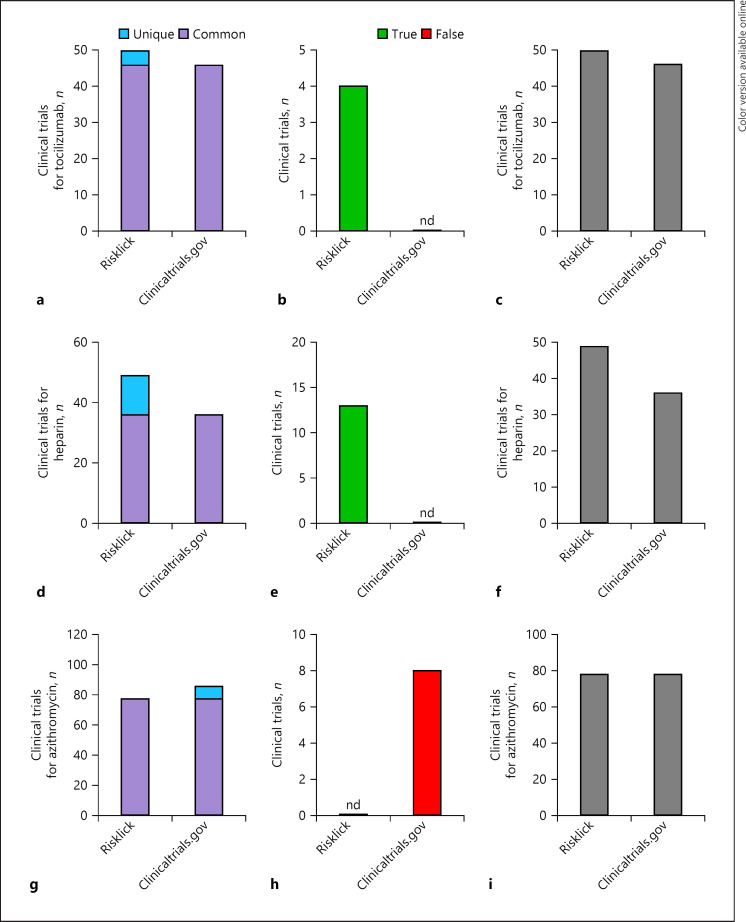
We further analyzed accuracy of both tools for drug classes. Risklick AI showed a higher number of relevant clinical trials for antibiotic (8.9%) (Fig. 2a–c), anticoagulant (29.4%) (Fig. 2d–f), and antiviral (47.2%) (Fig. 2g–i) drugs associated with COVID-19 in comparison with clinicaltrials.gov on the same reference database. Recall, precision, and F1 score measures for the 3 drug categories were systematically higher for Risklick AI compared to clinicaltrials.gov (online suppl. Table 3). The detailed analysis reveals that the higher score of Risklick AI is due to a higher number of true-positives (i.e., higher recall) and a lower number of false-positives (i.e., higher precision) in the unique findings cohort relative to clinicaltrials.gov (Fig. 2b, e, h). The analysis was then extended to specific drugs. Hydroxychloroquine (Fig. 1c–e), tocilizumab (Fig. 3a–c), and heparin (Fig. 3d–f) all presented a higher number of relevant clinical trials associated with COVID-19 compared to clinicaltrials.gov. Again, the higher score of Risklick AI is due to a higher number of true-positives and a lower number of false-positives unique findings in comparison with clinicaltrials.gov for these 3 drugs (Fig. 1d, 2b, e). Regarding azithromycin, the same number of relevant clinical trials was found in both search tools (Fig. 3g–i). However, in opposition to clinicaltrials.gov, Risklick AI uncovered no false-positive outcomes (Fig. 3h). Ultimately, no difference was observed between Risklick AI and clinicaltrials.gov regarding remdesivir (online suppl. Fig. 1). When taken together, Risklick AI presented an average recall of 99.25% compared to 86.61% for clinicaltrials.gov. By extension, Risklick AI also presented an F1 score of 97.59%, while clinicaltrials.gov had 88.57% (Table 1).
Fig. 2.
Risklick AI clinical trial search capacity for drug classes connected to COVID-19 compared to clinicaltrials.gov based on the same dataset. a−c Analysis of search capacity of registered clinical trials by Risklick AI and clinicaltrials.gov on the same database for antibiotic drugs. Clinical trials were separated between common and unique outcomes (a). Unique outcomes were validated and separated between true-positives (true) and false-positive (false) results (b). The final total number of true-positive clinical trials is the addition of common findings and unique, true-positive findings (c). The same procedure was performed for anticoagulant (d–f) and antiviral (g–i) drugs.
Fig. 3.
Risklick AI clinical trial search capacity for specific treatments associated with COVID-19 in comparison with clinicaltrials.gov on the same dataset. a–c Analysis of search capacity of registered clinical trials by Risklick AI and clinicaltrials.gov on the same dataset for tocilizumab. Clinical trials were separated between common and unique outcomes (a). Unique outcomes were validated and separated between true-positive (true) and false-positive (wrong) results (b). The total final number of true-positive clinical trials is the addition of common findings and unique, true-positive findings (c). The same procedure was performed for heparin (d–f) and azithromycin (g–i). nd, no data.
Table 1.
Risklick AI, clinicaltrials.gov, and PubMed average recall, precision, and F1 score
| Research tool | Recall, % | Precision, % | F1 score, % |
|---|---|---|---|
| Risklick | 99.25 | 96.07 | 97.59 |
| Clinicaltrials.gov | 86.61 | 91.43 | 88.57 |
| Risklick | 86.66 | 94.38 | 90.28 |
| PubMed | 61.26 | 88.22 | 71.63 |
Risklick AI Search Performance regarding COVID-19-Related Publications
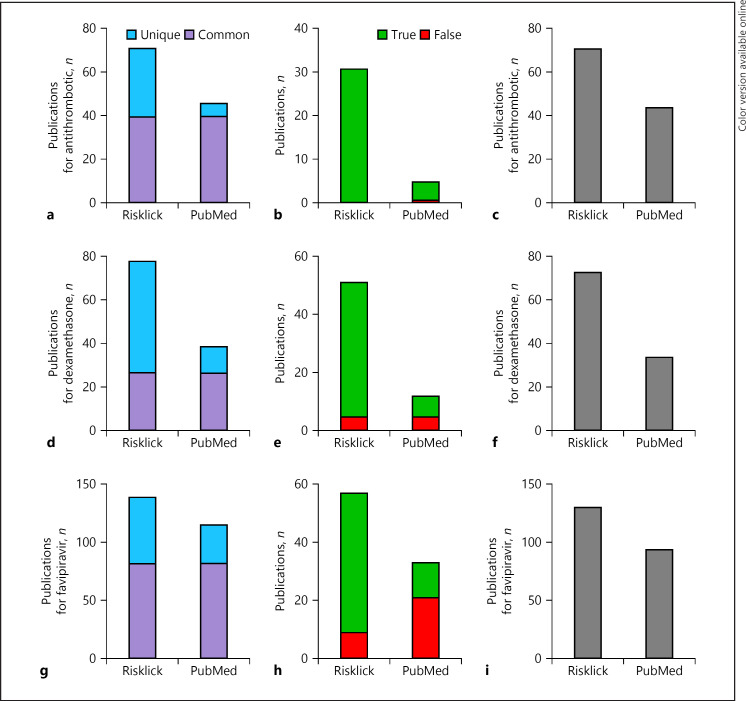
The data retrieval was extended to COVID-19-related scientific publications by comparing Risklick to PubMed search capacities. We restricted the search to the PubMed database using the Boolean search tool. We investigated the number of relevant publications restricted to COVID-19 for antithrombotic (+61.4%) (Fig. 4a–c), dexamethasone (+114.3%) (Fig. 4d–f), and favipiravir (+38.3%) (Fig. 4g–i). As for the comparison with clinicaltrials.gov, the superiority of Risklick AI compared to PubMed is due to a more important number of true-positives and a lower amount of false-positive unique findings (Fig. 4b, e, h). Taken together, the Risklick search presented an average recall of 86.66% compared to 61.26% for PubMed. In addition, the average F1 score for Risklick reached 90.28% compared to 71.68% for PubMed (Table 1).
Fig. 4.
Risklick AI publication search capacity for specific treatments associated with COVID-19 compared to PubMed on the same publication dataset. a–c Analysis of search capacity of COVID-19-related publications by Risklick AI and PubMed on the same publication dataset for antithrombotic. Publications were separated between common and unique outcomes (a). Unique outcomes were validated and separated between true-positive (true) and false-positive (wrong) results (b). The total final number of true-positive publications is the addition of common findings and unique, true-positive findings (c). The same procedure was performed for dexamethasone (d–f) and favipiravir (g–i).
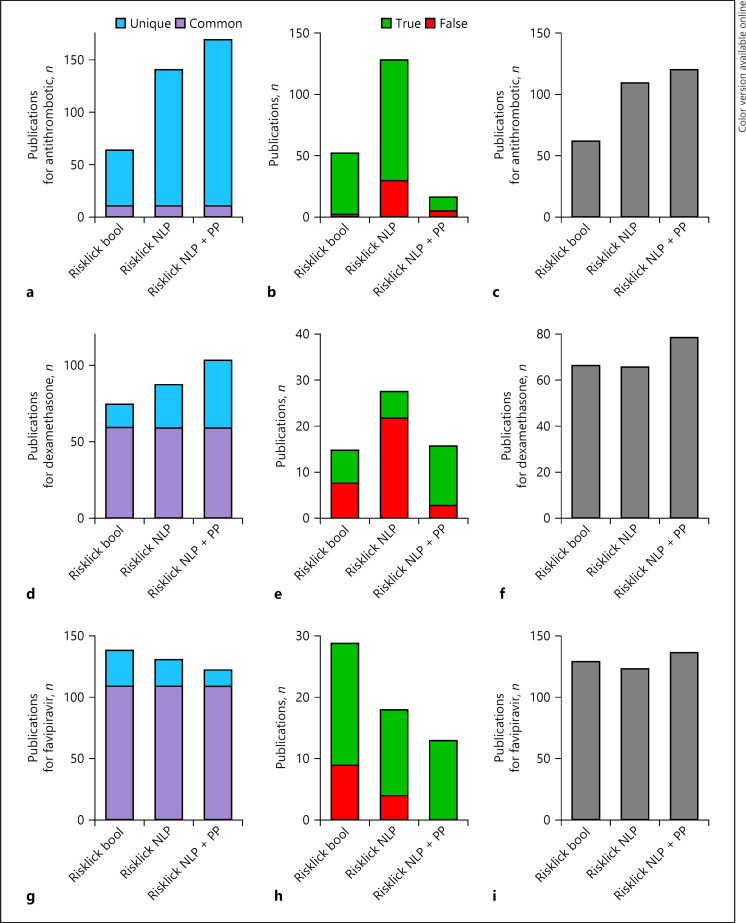
Evaluation of Risklick AI Publication Search Tool
Risklick AI offers the possibility to find COVID-19-related publications using Boolean-based search or NLP-based search methods and further combining the results of both methods. Here, we compare the capacity of each technology to find COVID-19-related publications. Hence, Boolean-based search (“Risklick bool”), NLP-based search (“Risklick NLP”), and NLP-based search supplemented with preprint (PP) publications (“Risklick NLP + PP”) were compared for antithrombotic (Fig. 5a–c), dexamethasone (Fig. 5d–f), and favipiravir (Fig. 5g–i). The searches in Risklick AI and clinicaltrials.gov are run based on the same dataset for the specific day and based on the same queries. Overall, Risklick AI NLP and Risklick NLP + PP offer more publications than Boolean-based search (+23.7% and +118.3%, respectively), although each search strategy presents various rates of false-positive outcomes (Fig. 5b, e, h). Used synchronously, both search methods offer a more complete, pertinent overview of currently available literature on the given treatments linked to COVID-19. Regarding clinical trials, clinicaltrials.gov uses Medical Subject Headings terms for query expansion, but does not match misspelled or differently spelled words for a disease or intervention. Risklick AI combines query expansion technology based on ontology defined by experts together with NLP techniques. The NLP techniques allow us to better deal with misspelled and similarly spelled words, which improved the quality of the search.
Fig. 5.
Risklick AI publication search capacity for specific treatments associated with COVID-19 using Boolean or NLP search methods. a–c Analysis of search capacity of COVID-19-related publications by Risklick AI using the Boolean search tool (bool), NLP research tool, and NLP with the database extended to PP publications for antithrombotic drugs. Publications were separated between common and unique outcomes (a). Unique outcomes were validated and separated between true-positive (true) and false-positive (wrong) results (b). The total final number of true-positive publications is the addition of common findings and unique, true-positive findings (c). The same procedure was performed for dexamethasone (d–f) and favipiravir (g–i). NLP, natural language processing; PP, preprint.
Discussion
The COVID-19 outbreak has resulted in one of the biggest waves of publications in the history of modern science [2, 23]. In these conditions, it has become clear that COVID-19 data retrieval and monitoring would be one of the main challenges of the current and future pandemics [24]. To address this dilemma, we automatically gathered and centralized all COVID-19 scientific information from scattered sources on a daily basis. Several intelligent algorithms and models were then developed to retrieve query relevant scientific evidences from a centralized database. Both Boolean and NLP-based search methods have been used to find query relevant scientific evidence.
In this study, the search performance of our methodology was compared to clinicaltrials.gov when screening the same database of clinical trials. Several molecules were selected to this purpose based on their connection to COVID-19 trials currently performed worldwide, as well as their important number of citations in the scientific literature. Overall, the abilities of the Risklick AI method to find relevant clinical trials against specific intervention queries were higher than the reference search tools, both for drug classes and for single treatments. Interestingly, the Risklick AI performance was largely due to a higher true-positive and lower false-positive outcome in comparison with clinicaltrials.gov. We believe this is due to the power of the full-text search engine combined with the Boolean model plus the improved semantics brought by the COVID ontology.
When extended to COVID-19-related publications, Risklick AI also confirmed a superior search capability compared to the medical reference tool PubMed, using the Boolean search engine. By extension, we compared the capacities of Risklick AI to find the scientific COVID-19 literature for pharmacological keywords using the Boolean and NLP approach. Molecules and categories selected for this analysis were chosen based on their relevance to COVID-19. These molecules were not engaged into numerous clinical trials as for molecules chosen in the clinicaltrials.gov comparison. We observed that both strategies offered a broad overview of key search articles with a high proportion of unique outcomes. In addition, we also confirmed the capacity of Risklick AI to find PP literature database with a high true-positive outcome, allowing for broad search perspectives in a context of permanent novelty not covered by PubMed.
On the one hand, Boolean search is still used in recent platforms like PubMed, Embase, and others. On the other hand, recent advancements in NLP and full-text searches enable better gathering of queries, sentences, and documents. These developments reduce the need for preprocessing and normalization steps, and they improve the quality of context-based searches.
Our methodology offers 2 search interfaces to find documents on the same datasets: one for Boolean search and one for NLP context-based search. This way users can arbitrarily combine the results of both approaches and thus improve precision and recall of their results. By extension, the evaluation results demonstrate the potential of the proposed method to help scientists and decision makers to triage key information out of the torrent of scientific studies from the COVID-19 pandemic. Consequently, Risklick AI could play a key role in the development of novel drugs and strategies targeting COVID-19 and could therefore become an important ally in fields such as pharmacology and epidemiology to organize the medical response against the SARS-CoV-2 virus. Moreover, in perspective of the current situation, Risklick AI could play a primordial role in the monitoring of all COVID-19 vaccines effectiveness, particularly in perspective of the numerous variants and associated serotypes of SARS-CoV-2. By extension, Risklick AI could offer significant advantages in the data management of other diseases and pathologies for clinicians and fundamental researchers. Since the underlying technology is generic, the framework can be used in other diseases and areas to manage relevant scientific evidences.
Statement of Ethics
This study did not involve human or animal material or data. Ethical approval was not required.
Conflict of Interest Statement
The authors Q.H., N.B., L.v.M., and P.A. are working for Risklick AG.
Funding Sources
This work was supported by the Innosuisse project, Funding No. 41013.1 IP-ICP.
Author Contributions
Q.H., P.A., and N.B. designed the study. Q.A., P.A., and N.B. wrote the manuscript. D.V.A., S.F., and D.T. designed and implemented the clinical trial and publication retrieval technologies. Q.H. and N.B. performed the experimental work. All authors had full access to the data, helped draft the report or critically revised the draft, contributed to data interpretation, reviewed, and approved the final version of the report.
References
- 1.Thorlund K, Dron L, Park J, Hsu G, Forrest JI, Mills EJ. A real-time dashboard of clinical trials for COVID-19. The Lancet Digit Health. 2020 Jun;2((6)):e286–7. doi: 10.1016/S2589-7500(20)30086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palayew A, Norgaard O, Safreed-Harmon K, Andersen TH, Rasmussen LN, Lazarus JV. Pandemic publishing poses a new COVID-19 challenge. Nat Hum Behav. 2020 Jul;4((7)):666–9. doi: 10.1038/s41562-020-0911-0. [DOI] [PubMed] [Google Scholar]
- 3.Brainard J. Scientists are drowning in COVID-19 papers. Can new tools keep them afloat? Science. 2020 May [Google Scholar]
- 4.Teixeira da Silva JA, Tsigaris P, Erfanmanesh M. Publishing volumes in major databases related to Covid-19. Scientometrics. 2020 [Online ahead of print] doi: 10.1007/s11192-020-03675-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besançon IL, Peiffer-Smadja IN, Segalas IC, Jiang H, Id PM, Smout IC, et al. Open science saves lives: lessons from the COVID-19 pandemic. BioRxiv. 2020 Aug; doi: 10.1186/s12874-021-01304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutson M. Artificial-intelligence tools aim to tame the coronavirus literature. Nature. 2020 Jun; doi: 10.1038/d41586-020-01733-7. [DOI] [PubMed] [Google Scholar]
- 7.Extance A. How AI technology can tame the scientific literature. Nature. 2018 Sep;561((7722)):273–4. doi: 10.1038/d41586-018-06617-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, Allot A, Lu Z. Keep up with the latest coronavirus research. Nature. 2020 Mar;579((7798)):193. doi: 10.1038/d41586-020-00694-1. [DOI] [PubMed] [Google Scholar]
- 9.Boutron I, Chaimani A, Meerpohl JJ, Hróbjartsson A, Devane D, Rada G, et al. The COVID-NMA project: building an evidence ecosystem for the COVID-19 pandemic. Ann Intern Med. 2020;173((12)):1015–7. doi: 10.7326/M20-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel J, James M. Speech and language processing. 2nd ed 2008. [Google Scholar]
- 11.Wolf T, Debut L, Sanh V, Chaumond J, Delangue C, Moi A, et al. HuggingFace's transformers: state-of-the-art natural language processing. ArXiv. 2019 Oct [Google Scholar]
- 12.Teodoro D, Mottin L, Gobeill J, Gaudinat A, Vachon T, Ruch P. Improving average ranking precision in user searches for biomedical research datasets. Oxford: Database; 2017. p. p. 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris ZS. Distributional structure. Word. 1954 Aug;10((2–3)):146–62. [Google Scholar]
- 14.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. ArXiv. 2018 Oct;1:4171–86. [Google Scholar]
- 15.Ferdowsi S, Borissov N, Kashani E, Alvarez DV, Copara J, Gouareb R, et al. Information retrieval in an infodemic: the case of COVID-19 publications. BioRxiv. 2021 Jan; doi: 10.2196/30161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tse T, Williams RJ, Zarin DA. Update on registration of clinical trials in clinical. Chest. 2009 Jul;136((1)):304–5. doi: 10.1378/chest.09-1219. trials.gov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahuja AS, Reddy VP, Marques O. Artificial intelligence and COVID-19: a multidisciplinary approach. Integr Med Res. 2020 Sep;9((3)):100434. doi: 10.1016/j.imr.2020.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WHO Clinical trials primary registries platform. https://www.who.int/clinical-trials-registry-platform/network/primary-registries.
- 19.Amati G, Van Rijsbergen CJ. Probabilistic models of information retrieval based on measuring the divergence from randomness. ACM Trans. Inf Syst. 2002 Oct;20((4)):357–89. [Google Scholar]
- 20.Diao L, Yan H, Li F, Song S, Lei G, Wang F. The research of query expansion based on medical terms reweighting in medical information retrieval. J Wireless Com Network. 2018 Dec;2018((1)):105. [Google Scholar]
- 21.COVID-19 projects of ISPM Available from: https://ispmbern.github.io/covid-19/living-review/collectingdata.html.
- 22.Hamel C, Kelly SE, Thavorn K, Rice DB, Wells GA, Hutton B. An evaluation of DistillerSR's machine learning-based prioritization tool for title/abstract screening − impact on reviewer-relevant outcomes. BMC Med Res Methodol. 2020 Dec;20((1)):256. doi: 10.1186/s12874-020-01129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callaway E. Will the pandemic permanently alter scientific publishing? Nature. 2020 Jun;582((7811)):167–8. doi: 10.1038/d41586-020-01520-4. [DOI] [PubMed] [Google Scholar]
- 24.Muhammad LJ, Islam MM, Usman SS, Ayon SI. Predictive data mining models for novel coronavirus (COVID-19) infected patients' recovery. SN Comput Sci. 2020 Jul;1((4)):206. doi: 10.1007/s42979-020-00216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]