Figure 1.
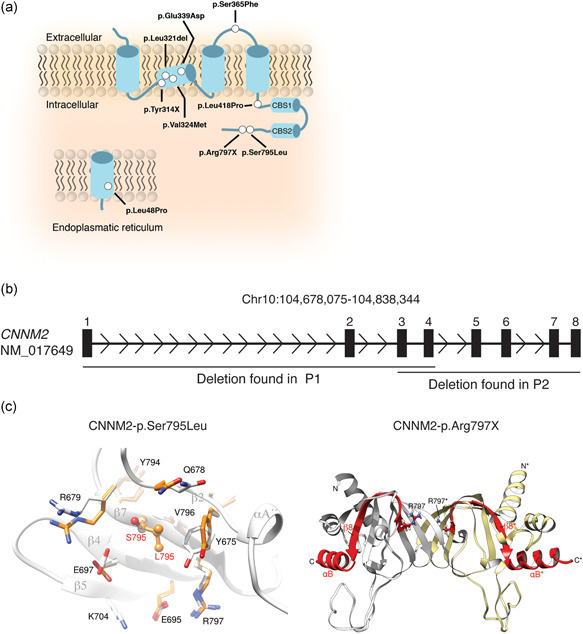
Identification of novel cyclin M2 (CNNM2) variants and their structural effects on the protein level. (a) Schematic overview of the localization of the mutations in the CNNM2 protein. Mutations are depicted as white dots. (b) Schematic overview of the genomic deletions found in the CNNM2 gene (h19; NM_017649) patients 1 and 2 (Table 1). Numbered boxes depict the exons within CNNM2. (c) The effect of mutation p.Ser795Leu was modeled using the crystal structure of CNNM2 (PDB code: 6DJ3) as a template. Left: Super composition of amino acid residues in the p.Ser795Leu environment in the native (light gray) and in the modeled mutation protein (orange). Substitution of serine to leucine (orange) at location 795 causes displacement of p.Glu697, Glu678, Arg679, and Tyr675, without affecting secondary elements (labeled in dark gray) significantly. Right: Crystal structure of the cNMP domain of CNNM2 (PDB code 6DJ3); the cNMP domain presents an α/β structure and self‐associates to form a tight elongated dimer (complementary subunits are colored in light gray and pale yellow, respectively), The β‐sheets occupy the central region and are sandwiched by α‐helices at both sides. The premature stop codon p.Arg797* disrupts the structure of strand β7 and causes the disappearance of the subsequent elements, strand β8 and helix αB (depicted in red) from the protein sequence. Arginine at position 797 is highlighted in balls and sticks. The long unstructured loop located in the bottom is visible only partially in one of the subunits in the crystals and has been modeled in the figure by applying the two‐fold symmetry that relates the two complementary subunits
