Fig. 1. The single nucleotide polymorphism c.454G>A in ZACN (rs2257020) and the resulting non-synonymous variation Thr128Ala in ZAC.
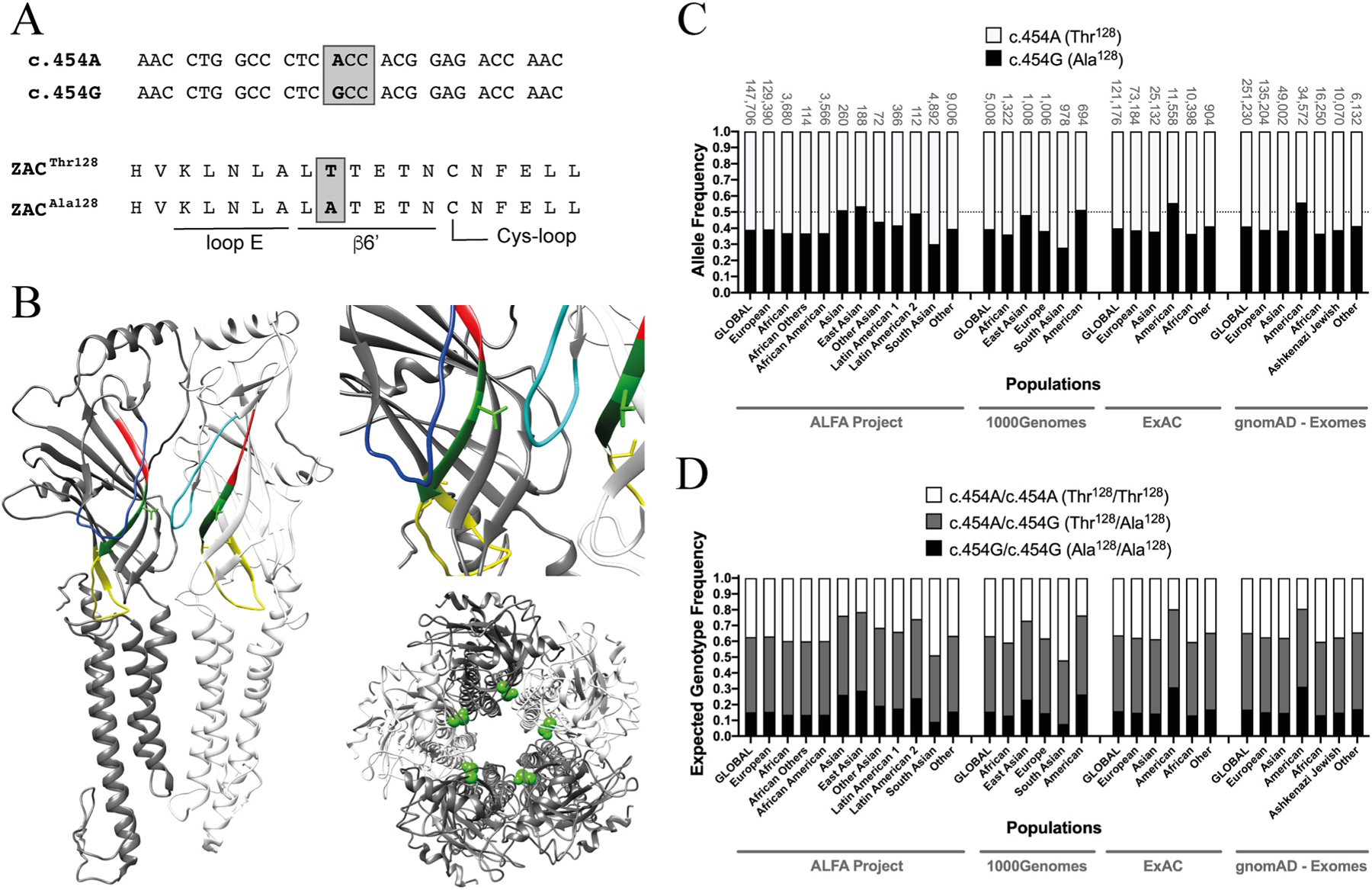
A. Alignments of the nucleotide sequence segments of the c.454A and c.454G alleles of ZACN (top) comprising the variation and of the amino acid sequence segments of the ZACThr128 and ZACAla128 proteins comprising the resulting Thr128Ala variation (bottom). The loop E and β6′ segments and part of the Cys-loop segment in ZAC are indicated. The c.454G>A SNP is given in bold, and the ACC and GCC codons encoding for Thr128 and Ala128 in ZAC, respectively, are boxed. B. Localisation of residue 128 (shown as Thr128) in a homology model of ZAC based on the cryo-EM structure of murine 5-HT3AR (PDB ID: 6HIN). Left: Two neighbouring subunits in the pentameric ZAC complex. The two Thr128 residues are shown in light-green stick, and the loop E, β6′ and Cys-loop segments are given in red, dark green and yellow, respectively. The β4-β5 loops are given in blue (dark-blue in the left subunit and light-blue in the right subunit). Right, top: A zoom in on Thr128 and its proximity in ZAC. Right, bottom: The ZAC complex viewed from an extracellular perspective with the Thr128 residues given in green spheres. C. Allele frequencies of the SNP c.454G>A in ZACN in various ethnic populations. The frequencies of the c.454A and c.454G alleles (encoding for ZACThr128 and ZACAla128, respectively) are represented by white and black in the bars, respectively. The sample sizes of the respective ethnic populations are given above each bar, and a line marker indicates the allele frequency 0.50. The data are from the Allele Frequency Aggregator (ALFA) Project (release version 20200227123210), the 1000 Genomes Project phase3 release V3 + (1000Genomes), the Exome Aggregation Consortium (ExAC) v0.3, and the Genome Aggregation Database (gnomAD - Exomes) and have been extracted from NCBI dbSNP (build 154, rs2257020) [29], where additional reports of allele frequencies for the SNP also can be found. D. Expected frequencies of the homozygous c.454A/c.454A (ZACThr128/ZACThr128), homozygous c.454G/c.454G (ZACAla128/ZACAla128) and heterozygous c.454A/c.454G (ZACThr128/ZACAla128) genotypes in different ethnic populations. The expected genotype frequencies were calculated from the selected allele frequency data for the SNP c.454G>A in ZACN (rs2257020) presented in Fig. 1C using the Punnett square under the assumption of the Hardy-Weinberg theorem of Mendelian genetics in the context of populations of diploid, sexually reproducing individuals.
