Abstract
Ras‐GTPase‐activating protein (GAP)‐binding protein 1 (G3BP1) is a multi‐functional protein that is best known for its role in the assembly and dynamics of stress granules. Recent studies have highlighted that G3BP1 also has other functions related to RNA metabolism. In the context of disease, G3BP1 has been therapeutically targeted in cancers because its over‐expression is correlated with proliferation of cancerous cells and metastasis. However, evidence suggests that G3BP1 is essential for neuronal development and possibly neuronal maintenance. In this review, we will examine the many functions that are carried out by G3BP1 in the context of neurons and speculate how these functions are critical to the progression of neurodegenerative diseases. Additionally, we will highlight the similarities and differences between G3BP1 and the closely related protein G3BP2, which is frequently overlooked. Although G3BP1 and G3BP2 have both been deemed important for stress granule assembly, their roles may differ in other cellular pathways, some of which are specific to the CNS, and presents an opportunity for further exploration.
Keywords: ALS, G3BP1, mRNA stability, neurodegenerative disease, RNA‐binding proteins, translation
G3BP1 (Ras‐GTPase‐activating protein (GAP)‐binding protein 1) is a multi‐functional protein involved in the assembly and dynamics of stress granules, and also other functions related to RNA metabolism. G3BP1 has been therapeutically targeted in cancers as its over‐expression is correlated with the proliferation of cancerous cells and metastasis. However, evidence suggests that G3BP1 is essential for neuronal development and possibly neuronal maintenance. The current Review examines the various functions of G3BP1 in the context of neurons, and we speculate how these functions are critical to the progression of neurodegenerative diseases. Additionally, we will highlight the similarities and differences, some of which are specific to the CNS, between G3BP1 and the closely related protein G3BP2, with a particular focus on stress granule assembly.
Abbreviations
- AD
Alzheimer's disease
- ADMA
assymetric dimethyl arginine
- AGD
argyrophilic grain disease
- ALS
amyotrophic lateral sclerosis
- ALS‐PDC
Gaumanian ALS‐parkinsonism‐dementia
- Caprin1
cytoplasmic activation/proliferation associated protein 1
- CBD
corticobasal degeneration
- ChAT
choline acetyltransferase
- circRNAs
circular RNAs
- CNS
central nervous system
- DDX
dead‐box RNA helicase
- FMRP
fragile X mental retardation protein
- FTD
frontotemporal dementia
- G3BP1
Ras‐GTPase‐activating protein (GAP)‐binding protein 1
- HD
Huntington’s disease
- HDAC6
histone deacetylase 6
- hnRNP
heterogeneous nuclear ribonucleoprotein
- IDRs
intrinsically disordered regions
- IRES
internal ribosomal entry sites
- JMJD6
Jumonji domain containing domain 6
- LLPS
liquid–liquid phase separation
- LTD
long‐term depression
- m6A
N6‐methyladenosine
- MAGE
melanoma antigen
- mGluR
metabotropic glutamate receptor
- MSP
multi‐system proteinopathy
- MSTD
multiple system tauopathy with dementia
- NPCs
neural precursor cells
- NTF2
nuclear transport factor 2
- PABP1
poly(A)‐binding protein 1
- PD
Parkinson’s disease
- PSP
progressive supranuclear palsy
- RBP
RNA‐binding protein
- RGG
arginine‐glycine‐glycine repeat
- RIN
rasputin
- RQC
ribosomal quality control
- RRM
RNA recognition motif
- SDMA
symmetric dimethyl arginine
- SIGMAR1
sigma non‐opioid intracellular receptor 1
- SRD
structure‐mediated decay
- SRE
Smaug recognition element
- TDP‐43
TAR DNA‐binding protein 43
- VZ
ventricular zone
- YB‐1
Y‐box‐binding protein 1
1. Introduction
Since its discovery in 1996 as a RAS‐binding partner, Ras‐GTPase‐activating protein (GAP)‐binding protein 1 (G3BP1) has been linked to a variety of biological processes, molecular functions and cellular compartments (Parker et al., 1996). Initially suggested to play a key role in the Ras signaling pathway via direct binding to Ras, more recent evidence contradicts this potential function (Annibaldi et al., 2011; Xu et al., 2013). Even if this initial function is debated, based on its role in development, RNA metabolism, degradation and stress response, G3BP1 seems to be a jack‐of‐all‐trades protein that is fundamental to cellular homeostasis.
Over the years, G3BP1 has been demonstrated to be essential for cell survival either as a key player to counter viral infections (Lloyd, 2016; White et al., 2007; White & Lloyd, 2011) or in cancer where it has been shown to aid tumors and cancer cells in overcoming chemotherapeutics and hostile conditions (Dou et al., 2016; Wang, Fu, et al., 2018; Zhang et al., 2015, 2019). However, to date, the exact molecular functions of G3BP1 in a physiological context have yet to be fully uncovered. In vivo evidence clearly indicates that G3BP1 is especially important for brain development and basal neuronal activities (Martin et al., 2013, 2016). The majority of what is known about G3BP1 is focused on its link to specialized RNA granules, termed stress granules, but largely in a non‐neuronal context.
The current review will describe the genetic and structural characteristics of G3BP1, as well as highlight important interactors. We will also discuss how G3BP1 has been demonstrated to be implicated in a number of key molecular mechanisms and how these may be associated with neurodegenerative disease. We will further explore important differences between G3BP1 and its paralogs G3BP2a and G3BP2b, and how they may be important in the CNS.
2. G3BP1 is not only important but essential for neuronal homeostasis
The exact functions of G3BP1 have been debated for many years. The importance of G3BP1 is highlighted by the generation of G3bp1−/− mice which exhibit late embryonic lethality (Zekri et al., 2005). Examination of G3bp1−/− neonates (on the 129/Sv mouse background) indicated that all organs were present and of normal size with the only marked abnormality being severe cell death in the brain (Zekri et al., 2005). Indeed, pyknosis and activated Caspase‐3 were prevalent within multiple brain regions, including CA1 pyramidal neurons of the hippocampus, cortical neurons and internal capsule neurons (Zekri et al., 2005). Importantly, this observation was absent in other organs, indicating that the lethality results from neuronal cell death in the CNS. Given that G3BP1 is ubiquitously expressed in both humans and mice, the finding that G3bp1 genetic deletion has a prominent and disproportionate effect on neurons was surprising and suggests that the encoded protein is essential to physiological neuronal functions.
In an effort to obtain viable pups, the original G3bp1−/− mice were subsequently crossed onto a 129/Balb/c background (Martin et al., 2013). Neurodegeneration was also observed in the brains of these mice, especially in the Purkinje layer of the cerebellum (Martin et al., 2013). While other CNS regions related to motor activity were not investigated, these mice displayed impaired motor coordination, dysfunctional synaptic transmission, and altered calcium homeostasis. Consolidation of long‐term depression was also impaired causing working memory deficits (Martin et al., 2016). In these mice, the apparent calcium homeostasis defect in the hippocampus was linked to neurocognitive defects and paralleled neurodegenerative diseases associated with aging in humans, such as Alzheimer's disease (AD) (Martin et al., 2013). In vitro studies in cultured hippocampal neurons showed that a combination of elevated metabotropic glutamate receptor stimulation and high basal calcium concentration led to an exacerbated internal release of calcium in these mice. With aging, this calcium misregulation could potentially be deleterious as this type of malfunction is linked to the progression of several neurological disorders including AD, Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), Huntington’s disease (HD), spinocerebellar ataxias, and stroke (Alavian et al., 2012; Bezprozvanny, 2009; Chung et al., 2015; Kawamoto et al., 2012; Mattson, 2007).
Taken together, these mouse models establish the physiological importance of G3BP1 to the CNS and suggest that dysfunction of the protein may be damaging for the brain.
3. Genetic and structural features of G3BP1
In humans, G3BP1 is encoded by the gene G3BP1 located on chromosome 5. Transcription gives rise to 14 different transcripts with only two encoding the 466 amino acid protein, and the others being degraded by non‐sense‐mediated decay (GTEx). The difference between the two protein‐encoding transcripts resides in the size of their untranslated regions. The longer transcript contains a 5’UTR of 132nt and a 3’UTR of 8,694nt, whereas the shorter transcript has a 5’UTR of 145nt and a 3’UTR of 1,263nt (GTEx, UCSC genome browser). These distinctions suggest a potential difference in the regulation and/or localization of the transcripts.
The G3BP1 protein has a theoretical molecular weight of 55kDa but runs at an apparent molecular weight of ~68KDa on standard Laemmli SDS‐PAGE. Structurally, G3BP1 is similar to proteins of the heterogeneous nuclear ribonucleoprotein (hnRNP) superfamily. The N‐terminal domain of the protein (amino acids 1‐139) has high structural similarity to nuclear transport factor 2 (NTF2), a small homodimer that plays an important role in nuclear transport via RanGDP import (Nehrbass & Blobel, 1996; Vognsen et al., 2013). Because of this similarity, the NTF2‐like domain of G3BP1 has been suggested to play a similar role in nuclear shuttling despite the fact that G3BP1 is primarily observed to be localized to the cytosol (Prigent et al., 2000; Vognsen et al., 2013). This domain is the most highly conserved region of the G3BP1 protein sequence and was previously suggested to be a RasGAP‐binding site, however, this has now been questioned (Annibaldi et al., 2011; Xu et al., 2013). The NTF2‐like domain is necessary for G3BP1 dimerization (Vognsen et al., 2013) and interaction with other proteins, such as cytoplasmic activation/proliferation‐associated protein 1 (Caprin1) (Kedersha et al., 2016). The region encoded by amino acids 139‐340 contains two intrinsically disordered regions (referred to as IDR1 and IDR2), the first half of which consists mainly of acidic amino acids (139‐221), whereas the second half is enriched for proline residues (PxxP; amino acids 222‐340). These regions are also associated with protein–protein interactions (Booker et al., 1993).
G3BP1 is an RNA‐binding protein (RBP), as indicated by the presence of an RNA recognition motif (RRM) located between amino acids 340‐415. This RRM has two conserved amino acid regions which can interact with RNA sequences of 2‐8 nucleotides via a binding platform formed by beta sheets (Cléry et al., 2008; Kennedy et al., 2002). G3BP1 also contains an arginine‐glycine‐glycine repeat (RGG) domain (amino acids 430‐461). This region harbors a third intrinsically disordered region (IDR3), which forms an undefined exposed structure because of RGG clusters that facilitate non‐specific RNA interactions. Post‐translational modifications in the RGG domain influences interactions with proteins and mRNA. For example, methylation in this domain increases steric hindrance and removes amino hydrogens that might be involved in hydrogen bonds and therefore modulate interactions with partner proteins (McBride & Silver, 2001). Since the RRM and RGG domains together form the G3BP1 RNA‐binding domain, it is plausible that post‐translational modifications can also modulate RNA interactions (Yang et al., 2020).
4. Regulation of G3BP1 mRNA
There are only a handful of studies focused on the regulation of G3BP1 mRNA. First, Y‐box‐binding protein 1 (YB‐1), a highly conserved cold shock domain family protein, regulates G3BP1 via binding to its 5’UTR to activate its translation (Somasekharan et al., 2015). YB‐1 has also been linked to neurodegeneration such that YB‐1 expression is neuroprotective in a mouse model of sporadic AD (Bobkova et al., 2015). Specifically, intranasal administration of YB‐1 into mice induced the formation of YB‐1 granules and positively correlated with a reduction in β‐amyloid plaques and showed an improvement in short‐term working memory (Bobkova et al., 2015). The authors noted that intact exogenous YB‐1 was detectable within cells for at least 2 hours, suggesting that it may be involved in other mechanisms regulating cell survival in conditions of AD‐type neurodegeneration (Bobkova et al., 2015).
MAGE‐B2 and DDX5 have recently been demonstrated to regulate G3BP1 transcripts in an antagonistic manner. MAGE‐B2, a protein member of the Melanoma antigen (MAGE) family, binds the G3BP1 mRNA 5’UTR to reduce its translation, whereas the Dead‐box RNA helicase DDX5 (or p68) binds to the G3BP1 5’UTR to increase translation (Lee et al., 2020). Interestingly, DDX5 binding to G3BP1 is inversely correlated with MAGE‐B2 binding and the deletion of the DDX5‐binding sequence in G3BP1 5’UTR inhibits MAGE‐B2 binding to G3BP1, suggesting that the two proteins compete for the same sequence (Lee et al., 2020). Moreover, this regulation was not dependent on YB‐1. Notably, the DDX5‐binding site is only present on the short G3BP1 transcript (Figure 1). Recent evidence indicates that G3BP1 transcripts are differentially expressed in different CNS regions, with the longer transcript being the predominant one expressed in the cerebellum compared to the frontal cortex (Sidibé et al., 2020). This implies that the relevance of this regulatory mechanism may vary within the CNS. Indeed, DDX5 malfunction has been suggested to be involved in tauopathies as its helicase activity is involved in the splicing of tau exon 10 (Kar et al., 2011). Misplicing of this exon leads to neurodegeneration because of an imbalance of the ratio of the tau4R/tau3R and is observed in progressive supranuclear palsy, corticobasal degeneration, multiple system tauopathy with dementia, argyrophilic grain disease, and even in some AD patients (Andreadis, 2006; Conrad et al., 2007; Gallo et al., 2007; Glatz et al., 2006; Umeda et al., 2004).
FIGURE 1.
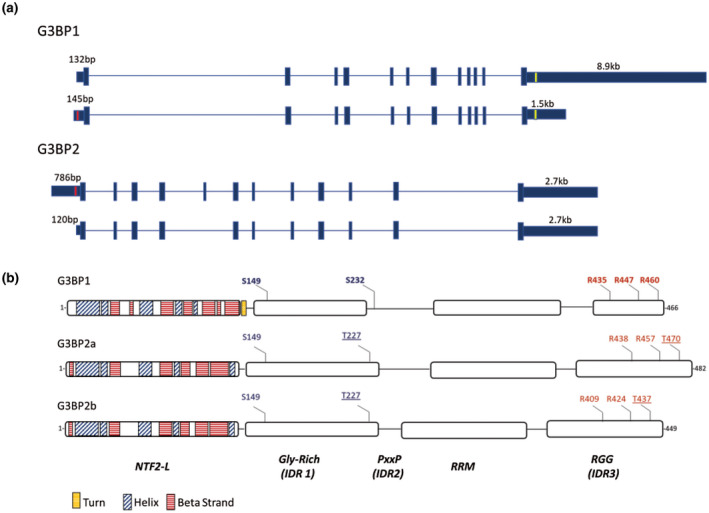
Sequence characteristics of G3BP proteins. (a) Schematic representation of G3BP1 and G3BP2 transcripts. G3BP regulating sequences are indicated (red: DDX5‐binding site, yellow: TAR DNA‐binding protein 43‐binding site). (b) Schematic representation of G3BP1, G3BP2a, and G3BP2b structural domains: nuclear transport factor 2 (NTF2)‐Like domain, Glycine rich‐domain (intrinsically disordered domain 1), PxxP (intrinsically disordered domain 2), RNA recognition motif (RNA‐recognition motif), and RGG (Arginine, Glycine rich domain, intrinsically disordered domain 3). Known sites of G3BP1 post‐translational modification are in bold (blue: phosphorylation; red: methylation). The three proteins were aligned and sites that aligned with the modified G3BP1 amino‐acids are displayed. Underline indicates sites that are not conserved.
G3BP1 is also regulated by TAR DNA‐binding protein 43 (TDP‐43), an RBP that is the primary constituent of the cytoplasmic inclusions observed in ALS, frontotemporal dementia (FTD), Gaumanian ALS‐parkinsonism‐dementia, and multi‐system proteinopathy (Forman et al., 2007; Kertesz, 2010; Scotter et al., 2015). It is notable that cytoplasmic inclusions of TDP‐43 are almost always associated with TDP‐43 nuclear depletion, suggesting TDP‐43 loss‐of‐function mechanisms may underlie these diseases. Indeed, TDP‐43 depletion results in decreased G3BP1 protein levels (McDonald et al., 2011) and recent work has uncovered that this is because of TDP‐43 binding and stabilizing the short G3BP1 transcript via a conserved 16‐nt element located within the 3’UTR (Sidibé et al., 2020). This work also demonstrated that G3BP1 mRNA levels are compromised in ALS/FTD neurons featuring TDP‐43 inclusions with concomitant nuclear depletion (Sidibé et al., 2020). Whether this is also true in other diseases featuring TDP‐43 pathology remains to be demonstrated.
5. What are the functions of G3BP1 and how are they linked to disease?
G3BP1 is most well‐known for its role in the nucleation of stress granules. In recent years, additional G3BP1 functions in axonal translation, ribosomal quality control (RQC), and transcriptional decay have been reported (Figure 2) (Fischer et al., 2020; Meyer et al., 2020; Sahoo et al., 2018). Overall, the recent literature suggests that G3BP1 is an essential regulator of cellular RNA metabolism.
FIGURE 2.
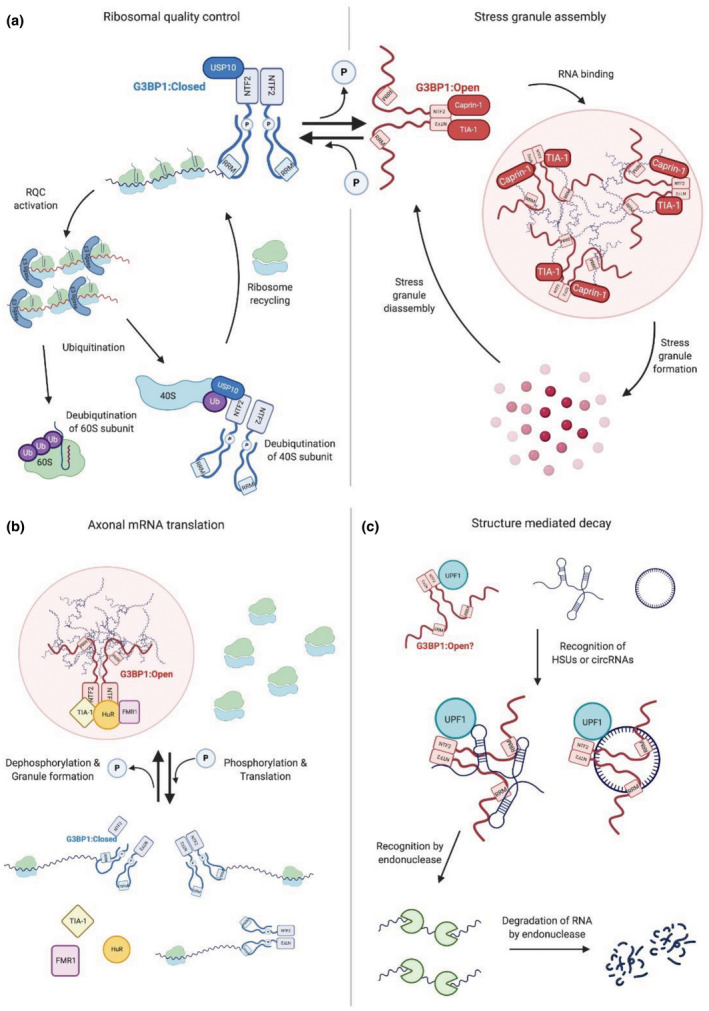
G3BP1 conformation affects its role in the cell. (a) G3BP1 functions in ribosomal quality control and stress granule assembly. In the closed conformation (blue), G3BP1 is phosphorylated and IDR1 acts as an autoinhibitory element to block the RNA recognition motif (RRM) from binding high concentrations of RNA. In the open conformation (red), G3BP1 is dephosphorylated, and the RRM binds RNA and can undergo liquid–liquid phase separation related to stress granule formation. (b) G3BP1 functions in axonal mRNA translation are illustrated. Specifically, G3BP1 is part of granules when the serines are dephosphorylated in combination with HuR, TIA‐1, and FMR1. When G3BP1 becomes phosphorylated, the granules disassemble and select mRNAs are translated in the axon. (c) G3BP1 function in structure‐mediated decay. (Figure created with BioRender.com)
5.1. G3BP1 and stress granules
Stress granules are cytoplasmic RNA‐protein condensates that form by liquid–liquid phase separation (LLPS) following exposure to adverse growth conditions (Guillén‐Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). G3BP1 was first shown to associate with stress granules in 2003, when it was co‐localized with HuR and TIA‐1 in granular structures following cellular exposure to sodium arsenite, an inorganic compound that can be used as a pesticide, antiseptic or carcinogen (Tourriere et al., 2003). Importantly, these puncta also contained polyadenylated mRNA as a result of interrupted translation because of eIF2α phosphorylation. Note, G3BP1 does not physically interact with eIF2α, whose phosphorylation leads to the rapid assembly of stress granules (Tourriere et al., 2003). Many other forms of external insults also lead to the formation of stress granules, but the proteomic composition as well as the assembly and dynamics of the granules vary according to stress exposure as well as cell type (Aulas et al., 2017). Regardless of the nature of the stress, G3BP1 has always been shown to be a part of these cytoplasmic biomolecular condensates.
5.1.1. G3BP1 in stress granule assembly
The ability for G3BP1 to mediate both protein–protein and protein–RNA interactions is essential to its role in stress granule assembly. Its NTF2‐like domain, RNA‐binding domain, and intrinsically disordered regions are important features to mediate the necessary protein and RNA interactions that drive the formation of these subcellular structures. Under external stress conditions, G3BP1 undergoes a conformational change where the dephosphorylation of S149 and S232 in IDR1 causes G3BP1 to adopt an open conformation which exposes the RRM and RGG domains, thereby enhancing G3BP1 dimer binding to RNA (Guillén‐Boixet et al., 2020; Yang et al., 2020). In basal conditions, IDR1 is phosphorylated at these sites and acts as an autoinhibitory element by blocking substantial RNA binding to the RRM and IDR3 in G3BP1 (Yang et al., 2020). Rather than bind specific sequences, G3BP1 preferentially binds RNAs that are either long, single‐stranded, or are prone to form an abundance of RNA–RNA interactions (Yang et al., 2020). In addition to RNA, many other proteins also aid in the formation of stress granules, whereas some proteins inhibit their formation. Using a model based on patchy colloid theories, the valency of a certain protein defines whether it will enhance or inhibit stress granule formation (Sanders et al., 2020). For example, G3BP1 and another stress granule nucleating protein UBAP2L each have large valencies, meaning they can mediate multiple interactions and therefore both greatly enhance the formation of stress granules. In addition, other G3BP1‐binding partners, such as Caprin1 and TIA‐1, decrease the critical concentration of RNA needed to bind G3BP1 and trigger LLPS. On the other hand, USP10, a deubiquitinase which can antagonize G3BP1‐Caprin1 interactions, has a low valency, so it acts as a cap and prevents granule formation by inhibiting additional RNA binding to G3BP1 and UBAP2L (Sanders et al., 2020). Taken together, the summation of the valencies of interactions defines the promotion or inhibition of granule assembly (Sanders et al., 2020). In the patchy colloid model, RNA acts as the substrate to promote the formation of the granules and increase valencies of these proteins (Sanders et al., 2020). RNA is the main component of the granules, and without RNA, granules will not form (Guillén‐Boixet et al., 2020). Importantly, the concentration of G3BP1 required for LLPS with RNA is quite low (65 mg/mL) compared to estimates of protein‐only condensates (400 mg/mL) (Guillén‐Boixet et al., 2020). Lastly, the binding of G3BP1 to RNA, in the form of condensates, is thought to inhibit RNA entanglement, which could otherwise lead to toxicity (Guillén‐Boixet et al., 2020). Given that G3BP1 regulates the formation and dynamics of stress granules, it has recently been proposed to be the central regulator of stress granule assembly (Guillén‐Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). In fact, reconstruction of a G3BP1‐like protein with all the appropriate structural features can effectively rescue stress granule assembly in G3BP depleted cells, although the protein and RNA composition of these reconstituted granules is likely very different (Yang et al., 2020).
Another model that has been proposed for stress granule assembly is centered on the concept of free mRNA (Bounedjah et al., 2014). Under physiological conditions, there is a higher proportion of mRNA bound to polysomes (polysomal mRNA) compared to non‐polysomal mRNA found in microscopic RNPs (Arava et al., 2003). In the presence of an external stressor, polysome disassembly results in a large increase in non‐polysomal mRNA in the cytoplasm. The mRNAs “freed” from protecting ribosomes may then seed the formation of mRNA‐rich granules because of the intrinsic tendency of RBPs to oligomerize (Bounedjah et al., 2014). In other words, this model proposes that excess non‐polysomal mRNA could be the reason for aggregation of self‐attracting RBPs, which leads to stress granule formation. In this mechanism, G3BP1 would bind non‐polysomal mRNA. Post‐translational modifications impacting the valency of G3BP1 could induce conversion of the protein structure, enhancing binding properties to RNA. In this case, SGs assembly would be primarily driven by mRNA interactions and secondarily by protein/protein interactions.
5.1.2. G3BP1 and stress granule composition
The RNA composition of stress granules has not been associated with a specific biological process, but is mainly composed of mRNAs with long‐coding sequences and poor translatability (Khong & Parker, 2018). The recruitment of RNA to stress granules is largely based on RNA structure and interaction potential. The influence of G3BP1 on the stress granule transcriptome is thought to be minimal (Matheny et al., 2020). One common mRNA modification that has been associated with stress granule targeting is the N6‐methyladenosine (m6A) modification. m6A‐modified RNAs have an enhanced ability to phase separate, and this tracks positively with a report that m6A‐modified RNAs are recruited to stress granules by interacting with the YTHDF family of proteins (Ries et al., 2019). YTHDF proteins are components of stress granules and are thought to promote stress granule formation (Fu & Zhuang, 2020). However, using a systematic mass spectrometry‐based screening of m6A‐modified mRNA interactors, G3BP1 was found to repel RNA molecules with m6A sites (Edupuganti et al., 2017). While it is possible that G3BP1 and m6A‐modified mRNAs can co‐exist in stress granules, there is some debate on whether the m6A modification effectively targets mRNAs to these condensates.
If and how G3BP1 affects the protein composition of stress granules remains unclear. It has been shown that siRNA‐mediated knockdown of G3BP1 results in smaller stress granules and that this correlates with reduced mRNA steady‐state levels, suggesting that mRNA is insufficiently protected/more rapidly degraded (Aulas et al., 2015). Using super‐resolution microscopy, stress granules have been proposed to consist of a relatively stable inner core with a more dynamic outer shell where proteins and RNAs are continuously exchanged with the surrounding cytoplasm (Jain et al., 2016). This model positioned G3BP1 as an important component of stress granule cores, along with other well‐known stress granule proteins such as TIA‐1, Caprin1, and TIAR. However, considering new information on valencies (Sanders et al., 2020), the loss of G3BP1 could result in an overall decreased valency and therefore contribute to the outcome of stress granules having a smaller size. Indeed, this is observed when G3BP1 is depleted in cells, with these smaller granules showing fewer interactions with P‐bodies (Aulas et al., 2012, 2015). Because G3BP1 mediates many protein–protein interactions in stress granules via its NTF2‐like domain, one could speculate that the proteome of stress granules is severely affected by the loss of G3BP1. However, this question has not been directly assessed.
5.1.3. Post‐translational modifications of G3BP1 affect stress granule assembly
G3BP1 is subjected to numerous post‐translational modifications including phosphorylation, acetylation, methylation, and parylation (Table 1) (Bikkavilli & Malbon, 2011; Gal et al., 2019; Leung et al., 2011; Solomon et al., 2007). Each modification can individually or in concert influence G3BP1 functions. For example, specific arginine residues in the RGG domain of G3BP1 are differentially methylated by protein arginine methyltransferases 1 and 5 (PRMT1 and PRMT5). The knockdown of these methyltransferases or expression of a methylation‐deficient G3BP1 mutant promotes the formation of stress granules upon stress exposure (Tsai et al., 2016). Mechanistically, PRMT1 methylates R435 and R447, whereas R460 is methylated by PRMT5. PRMT1‐mediated methylation of R447 strongly inhibits stress granule formation, and R435 and R460 remain methylated during stress granule formation (Tsai et al., 2016). The authors suggest that PRMT5 does not enter stress granules whereas PRMT1 does and promotes R447 methylation in situ, stimulating stress granule disassembly by depleting G3BP1 from the granule. This arginine demethylation requirement is altered with different types of stress. For example, assymetric dimethyl arginines (ADMAs) are decreased on G3BP1 in arsenite stress, but symmetric dimethyl arginines are decreased with heat stress and thapsigargin‐induced ER stress (Tsai et al., 2016). It is noted that chemical inhibition of PRMT1 and PRMT5 results in a higher proportion of smaller stress granules, similar to what is observed in G3BP1‐depleted conditions (Aulas et al., 2012; McDonald et al., 2011; Tsai et al., 2016).
TABLE 1.
Summary of G3BP1 post‐translational modifications and their effects on the protein
| PTM | Residues | Proteins involved | Effects | References |
|---|---|---|---|---|
| Acetylation | K376 | CBP/p300 |
Prevents G3BP1 binding to mRNA Prevents mRNA‐dependent binding to PABP1 SG disassembly |
Gal et al., (2019) |
| Deacetylation | K376 | HDAC6 | Required for SG formation | |
| Methylation | R435 | PRMT1 | Methylated during SG assembly and maintenance | Tsai et al., (2017) |
| R447 | PRMT1 | Inhibits stress granules formation | ||
| R460 | PRMT5 | Methylated during SG assembly and maintenance | ||
|
R435 a R447 a |
PRMT8 | Spine maturation | Lo et al., (2020) | |
| Demethylation |
R435 R447 R460 |
JMJD6 | Promotes SG formation and G3BP1 oligomerization | Tsai et al., (2017) |
| Phosphorylation | S149 | Casein kinase 2 |
Blocks G3BP1 RNA binding Allows axonal regeneration |
Sahoo et al., (2018), Sahoo et al., (2020) and Yang et al., (2020) |
| S232 | — | |||
| Dephosphorylation | S149 | — |
Induces G3BP1 open conformation Enhances G3BP1 dimer binding to RNA |
Guillén‐Boixet et al., (2020) and Yang et al., (2020) |
| S232 | — | |||
| ADMAs (asymmetric dimethyl arginine) | — | — | Decreased in SA stress | Tsai et al., (2017) |
| SDMAs (symmetric dimethyl arginine) | — | — | Increased in HS and ER stress | |
| Parylation | — | PARP | Facilitates RBP recruitment to SGs | Leung et al., (2011) |
Abbreviations: ADMA, assymetric dimethyl arginine; G3BP1, Ras‐GTPase‐activating protein (GAP)‐binding protein 1; HDAC6, histone deacetylase 6; PABP1, poly(A)‐binding protein 1; RBP, RNA‐binding protein; SDMA, symmetric dimethyl arginine.
Lo et al, (2020) reported as R433 and R445 of the mouse sequence corresponding to R435 and R447 of the human sequence.
In neurons, G3BP1 is also dimethylated by PRMT8 (Bikkavilli & Malbon, 2011; Guo et al., 2014). PRMT8 expression is highly restricted to the CNS with spinal cord motor neurons exhibiting a high abundance of ADMAs because of PRMT8 (Kousaka et al., 2009; Simandi et al., 2018; Taneda et al., 2007). The presence of this post‐translational modification provides protection against environmental stress (Simandi et al., 2018). Notably, ADMA levels decline during embryonic development in the mouse, however, choline acetyltransferase positive motor neurons selectively maintain high ADMA levels in the adult spinal cord (Simandi et al., 2018). In mice, persistent stress caused by the absence of PRMT8 activity decreases activation of pro‐survival and regeneration pathways, even under aging‐related oxidative and ER stress, ultimately leading to a progressive loss of muscle strength because of premature destabilization of neuromuscular junctions (Simandi et al., 2018). Moreover, PRMT8 expression is increased in the hippocampus during spine maturation and depletion of PRMT8 impairs spine maturation and alters actin dynamics; importantly, these phenotypes are mimicked by G3BP1 depletion (Lo et al., 2020). Thus, defects in the motor neuron‐specific methyltransferase PRMT8 induce G3BP1 dysfunction, developmental problems, and accelerated aging that could potentially be rescued by G3BP1 activity. Interestingly, this activity can be countered by the stress granule resident protein Jumonji domain‐containing domain 6 which directly interacts with and reduces G3BP1 monomethylation, thereby promoting G3BP1 oligomerization and stress granule formation (Tsai et al., 2017).
Upon stress, G3BP1 also exhibits a significant increase in parylation (polyADP‐ribosylation), a post‐translational modification resulting from the addition of a macromolecular polymer of 2‐200 ADP‐ribose subunits, organized in linear or branched chains (Schreiber et al., 2006). The formation of G3BP1 positive stress granules is pADPr‐dependent and helps to facilitate the recruitment of other stress granule RBPs (Chang et al., 2009; Duan et al., 2019; Kotova et al., 2009). Indeed, this modification is proposed to act as a scaffold for protein recruitment at stress granules, or other cellular structures, consistent with G3BP1’s function as a stress granule nucleator (Duan et al., 2019; Isabelle et al., 2012).
Lastly, it is reported that an acetylation event impairs G3BP1’s RNA‐dependent interaction with poly(A)‐binding protein 1 and mRNA, thus promoting G3BP1 disaggregation and consequent stress granule disassembly (Gal et al., 2019). G3BP1 acetylation is regulated by histone deacetylase 6 (HDAC6) and CBP (cyclic AMP response element‐binding protein) and its homolog p300 (Gal et al., 2019). HDAC6 activity is reportedly required for the formation of stress granules (Kwon et al., 2007). Specifically, HDAC6 interacts with G3BP1 and although this interaction is independent of HDAC6 activity, it is modulated by G3BP1 phosphorylation at S149. Similar to G3BP1 knockdown conditions, inhibition of HDAC6 deacetylation activity yields smaller arsenite‐induced stress granules compared to untreated control cells (Kwon et al., 2007). While HDAC6 is known to have pleiomorphic effects, it is noteworthy that HDAC6 down‐regulation is observed in several neurodegenerative diseases including AD, PD, and HD, and HDAC6 is regulated by TDP‐43 (Fiesel et al., 2010; Simões‐Pires et al., 2013).
5.1.4. Stress granules in neurodegenerative disease
Mutations in many stress granule components have been linked to neurodegenerative diseases (Wolozin & Ivanov, 2019) and many others have been predicted to be involved in these neurodegenerative diseases (Bakkar et al., 2018). However, it is important to note that many follow‐up studies focused on stress granule dynamics have been performed in immortalized cell lines (e.g. HeLa, U2OS, HEK293) or used over‐expression to express proteins that are prone to undergo phase separation (Aulas & Vande Velde, 2015). The contribution of stress granule dynamics in neurons and/or in vivo is important to deepen our understanding of how these condensates are affected in neurodegenerative diseases. In general, there are three ways that have been proposed in which stress granules could be associated with neurodegeneration (Li et al., 2013). First, stress granules could fail to assemble properly and therefore are not functional, resulting in increased neuronal vulnerability (Wolozin & Ivanov, 2019). Since stress granules are associated with mRNA protection, improper assembly would lead to an increase in non‐translating RNA, which could have various detrimental effects associated with dysregulated RNA metabolism. (Aulas et al., 2015). Second, stress granules form during times of stress but do not disassemble, and over time evolve from a liquid phase to a solid phase, effectively seeding the cytosolic inclusions frequently associated with neurodegenerative diseases (Baron et al., 2013). Lastly, stress granules that fail to disassemble could sequester important translation factors, transport proteins, or other crucial cellular chaperones (Wolozin & Ivanov, 2019). Taken together, these hypotheses suggest that stress granule dysfunction in neurodegeneration is likely mediated by a defective balance between RNA‐binding proteins and RNAs crucial for LLPS (Sidibé & Vande Velde, 2019; Wolozin & Ivanov, 2019). It is likely that a combination of these processes contribute to the onset and progression of neurodegeneration, and therefore understanding how stress granules behave in neurons and the functional consequences of their dysfunction is essential. Given that G3BP1 is the central regulator of stress granules, studying it in the context of neurodegenerative disease will likely help elucidate the nature of the defect.
5.2. G3BP1 in RNA and ribosomal stability
The in‐depth understanding we have of G3BP1 in the assembly of stress granules opens an interesting question: what is the function of G3BP1 in steady‐state conditions when the cell is not being challenged by environmental stress? The literature suggests that G3BP1 depletion impacts the stress granule proteome more so than its associated transcriptome (Edupuganti et al., 2017; Markmiller et al., 2018). Therefore, one could infer that the RNA‐binding capacity of G3BP1 is important in steady‐state conditions. The G3BP1 ortholog in Drosophila, Rasputin (RIN) was originally linked to cell differentiation but has recently been shown to be associated with polysomes to promote the translation of short, highly translated transcripts (Laver et al., 2020). Similar to mammalian cell lines, RIN interacts with Caprin1 and FMR1 in Drosophila early embryos. Transcriptomic analyses indicate that RIN is associated with many abundant transcripts that are linked to oxidative phosphorylation, the ribosome, and transcriptional control. Importantly, these transcripts were depleted for Smaug recognition elements (SRE), which are elements associated with mRNA destabilization and therefore decreased translation (Chen et al., 2014). Lastly, RIN‐bound transcripts were neither enriched nor depleted for m6A‐modifications compared to the general cellular level (Laver et al., 2020). The authors proposed that during extracellular stress, the association of G3BP1 with stress granules is two‐pronged: (a) it functions to stabilize vulnerable mRNAs; and (b) it directly lowers translation levels since it is no longer directly bound to the ribosome. In a study using human PC‐3 cells, the authors compared the transcripts bound by either polysomes or G3BP1 following arsenite stress (Somasekharan et al., 2020). Interestingly, transcripts that bind G3BP1, but not polysomes after stress, are associated with pathways related to oxidative phosphorylation and mitochondrial transport (Somasekharan et al., 2020), similar to what was found with RIN (Laver et al., 2020). Transcripts that were polysome‐enriched and G3BP1‐depleted during stress were associated with cell division and DNA repair (Somasekharan et al., 2020). Finally, transcripts that were both polysome and G3BP1‐enriched during stress were involved in stress response pathways (Somasekharan et al., 2020). Overall, this could indicate that G3BP1 also plays an important regulatory role in translation in times of stress.
Transcript analyses in the mouse brain suggest that G3BP1 also binds and stabilizes transcripts with retained introns (Martin et al., 2016). Specifically, G3BP1 preferentially binds to non‐coding sequences of RNA and increases the stability of transcripts with retained intronic sequences as well as long non‐coding RNAs (Martin et al., 2016). Indeed, G3BP1 depletion was associated with a decrease in intron‐retained transcripts, leading to widespread changes in the stability of premature RNAs in the cerebellum (Martin et al., 2016). Among the enriched transcripts, genes involved in synaptic transmission, especially glutamate‐related, were over‐represented. Ultimately, G3BP1‐regulated transcriptional changes induced ataxia, exacerbated neuronal responses and impairment of hippocampal plasticity (Martin et al., 2016). The role of G3BP1 in the stability of short, highly translated transcripts (Laver et al., 2020; Somasekharan et al., 2020) compared to its role in the stability of transcripts with retained introns (Martin et al., 2016) could suggest a difference in its RNA stability function in dividing cells and neurons.
In addition to being associated with stabilizing specific mRNAs, G3BP1 has been suggested to play an important role in the stabilization of the ribosome itself (Meyer et al., 2020). The RQC pathway is activated during the translation of aberrant mRNAs and the arrested complex is dissociated into 40S and 60S subunits, followed by aberrant mRNA decay (Pisareva et al., 2011; Shoemaker et al., 2010). Recycling of the 60S ribosomal subunit involves polyubiqutination of the nascent polypeptide by NEMF and LTN1 (Joazeiro, 2019). To recycle the 40S ribosomal subunit, USP10 and G3BP1 are directly involved in its deubiquitination to prevent its degradation (Meyer et al., 2020). As the need for recycling ribosomal subunits is a regular process, this result is in agreement with the fact that in a G3BP1 knockdown context, there is an overall increase in cellular ubiquitination (Anisimov et al., 2019) and decreased protein synthesis (Aulas et al., 2015). As many ribosomal subunits are components of stress granules (Khong et al., 2017), it would follow that G3BP1 is also present during the deubiquinatination of these proteins by USP10. In agreement with this, the C‐terminus of G3BP1 has been shown to interact with two 40S ribosomal proteins, RPS6 and RPS23, which were originally proposed to be important for stress granule assembly (Kedersha et al., 2016). However, replacing the C‐terminus of G3BP1 with heterologous sequences from other RBPs is sufficient to assemble SG‐like structures in G3BP1/2 double knockout cells (Yang et al., 2020), indicating that the interaction between G3BP1 and the ribosomal subunits is not necessary for this process. Given that the interaction between G3BP1 and USP10 inhibits stress granule formation, this function is anticipated to be relevant to unstressed/physiological conditions, where translation rates are higher (Kedersha et al., 2016). In conditions with low G3BP1 expression, ubiquitination is also increased for α‐synuclein and CFTRΔ508, two disease‐causing and aggregation‐prone proteins (Anisimov et al., 2019). Moreover, the increased ubiquitination of these proteins is related to increased aggregation and cell toxicity. Lastly, this study reported that decreased G3BP1 expression is associated with proteasome inhibition (Anisimov et al., 2019). Therefore, in the absence of G3BP1, ubiquitinated proteins accumulate and cannot be degraded, presumably increasing their toxicity to cells. This is potentially relevant to the many examples of ubiquitinated aggregates that accumulate in neurodegenerative disease‐affected neurons (Wolozin & Ivanov, 2019).
5.3. G3BP1 and structure‐mediated RNA decay
A recent study demonstrates that G3BP1 acts in concert with UPF1 to selectively degrade highly structured RNAs (Fischer et al., 2020). This mechanism, referred to as structure‐mediated decay (SRD), involves UPF1 and G3BP1 interaction in an RNA‐dependent manner at highly base‐paired regions. The binding of the helicase UPF1 to these regions permits the unwinding of the double‐stranded RNA regions and facilitates G3BP1 binding, leading to the degradation of the targets via an unknown endonuclease, which is hypothesized to be G3BP1 (Fischer et al., 2020). Note, G3BP1 has been previously reported to bind highly structured sequences such as viral internal ribosomal entry sites in order to inhibit translation (Galan et al., 2017).
To date, SRD has been demonstrated to have two main functions. First, to differentially regulate specific isoforms of mRNAs, a function of notable importance in neurons since long and highly structured transcripts are more prominent in this cell type (Bernat & Disney, 2015; Middleton et al., 2019). The complex structures facilitate the recognition of these transcripts by transporters and regulators for trafficking to axons and dendrites (Je et al., 2018; Middleton et al., 2019; Tushev et al., 2018). Second, SRD regulates circular RNAs, the levels of which change throughout development and aging, and are tightly regulated. Notably, these non‐coding RNAs can serve as sponges for miRNAs, as decoys to sequester RNA‐binding proteins, and can even be translated to protein (Yu & Kuo, 2019). In a neuronal context, these RNAs can be detrimental for cells and may drive neurodegeneration (Memczak et al., 2013; Piwecka et al., 2017; Wang, Liu, et al., 2018). For example, circHIPK2 functions as a sponge to sequester miR‐124 and inhibit its activity, resulting in increased sigma non‐opioid intracellular receptor 1 expression, a protein mutated and/or misregulated in several neurodegenerative diseases, including ALS and AD (Fehér et al., 2012; Gregianin et al., 2016; Luty et al., 2010; Maruszak et al., 2007; Mori et al., 2012; Tagashira et al., 2014; Ververis et al., 2020). Also, miR‐124 is a neuron‐specific microRNA that induces a switch from general to neuron‐specific alternative splicing, meaning that the loss of miR‐124 via the misregulation of circRNAs may impair neuronal development and renewal (Makeyev et al., 2007).
RNAs targeted for SRD are excluded from stress granules, thus providing G3BP1 an opportunity to regulate a distinct subset of RNAs and broadly influence the transcriptome (Fischer et al., 2020). Interestingly, UPF1 localizes to G3BP1‐positive stress granules (Brown et al., 2011). Delayed stress granule disassembly, as is reported in cells expressing ALS‐associated FUS or TDP‐43 mutations, may sequester the two proteins away from their targets and impair RNA metabolism post‐stress (Bosco et al., 2010; Gal et al., 2010). It has been suggested that G3BP1 and UPF1 could be subject to post‐translational modifications that serve to switch their function to either SRD or stress granules. Thus, impairments in the post‐translational modification code of G3BP1/UPF1 could be deleterious for cells (Fischer et al., 2020).
UPF1 is expressed at low levels but is tightly regulated for optimal cellular function such that elevated UPF1 levels leads to translational inhibition, whereas diminished levels result in RNA‐mediated toxicity (Barmada et al., 2015). UPF1 has already been associated with neurodegenerative disorders. Specifically, UPF1 expression is neuroprotective in mutant TDP‐43 and FUS ALS models; however, this does not extend to mutant SOD1‐mediated neurodegeneration (nor HD models) (Barmada et al., 2015), reinforcing the concept that mutant SOD1‐associated ALS occurs through mechanisms that are fundamentally dissimilar to those involving TDP‐43 and FUS. TDP‐43 and FUS bind to and trigger the alternative splicing of thousands of transcripts resulting in transcripts that are substrates for NMD and the formation of splicing by‐products such as circRNA (Lagier‐Tourenne et al., 2012; Zhou et al., 2013). Human UPF1 effectively rescues neurons from TDP‐43 over‐expression but fails to improve survival in TDP‐43 knockdown neurons. The authors suggest that the loss of endogenous TDP‐43 is not responsible for the toxicity associated with TDP‐43 over‐expression and that cell death resulting from the absence or overabundance of TDP‐43 is via distinct mechanisms (Barmada et al., 2015). Given that TDP‐43 regulates G3BP1 (Sidibé et al., 2020), it is plausible that in the context of TDP‐43 nuclear depletion, the lack of G3BP1 impedes UPF1 from carrying out its SRD‐related functions, resulting in toxicity. Moreover, FUS mutant proteins disturb stress granule disassembly such that G3BP1 remains in the granules longer than intended (Baron et al., 2013). Therefore, in the context of TDP‐43 and FUS pathology, G3BP1 may not be available to function in SRD, resulting in RNA‐related toxicity.
5.4. Neuron‐specific functions of G3BP1
5.4.1. G3BP1 and axonal growth
Several studies have noted the importance of axonal mRNA translation for neuronal growth, repair, and homeostasis (Klim et al., 2019; Sahoo et al., 2018, 2020; Sephton et al., 2014; Welshhans & Bassell, 2011; Willis et al., 2005). A number of SG proteins have been associated with axonal translational, including G3BP1. Specifically, G3BP1 assembles into axonal SG‐like structures that are approximately the size of the SG cores proposed by Jain et al (Sahoo et al., 2018). As a part of these granules, G3BP1 modulates protein synthesis and axon growth. In response to axotomy, a rapid increase in these aggregates is observed in the axon. Disaggregation of these granules is triggered by G3BP1 phosphorylation at S149 by the calcium‐dependent activation of Casein Kinase 2 and correlates with accelerated regeneration (Sahoo et al., 2018, 2020). This is in agreement with the recent report that phosphorylation of G3BP1 at this site promotes the closed conformation state which defavors LLPS (Yang et al., 2020). Naive and injured neurons have different protein needs in order to respond to their different growth states. For example, Nrn1 translation promotes neurite growth and axonal growth, whereas Impβ1 protein is retrogradely transported to activate regeneration‐associated gene expression in the soma and likely decreases axon elongation because of its role in axon length sensing (Sahoo et al., 2018). Both Nrn1 and Impβ1 mRNAs were released from G3BP1 axonal SG‐like structures (Sahoo et al., 2018). In sum, axonal G3BP1 is a specific modulator of intra‐axonal protein synthesis and axon growth. The fact that G3BP1 is aggregated in uninjured axons points toward unknown functions for G3BP1 in physiological conditions. Moreover, these axotomy experiments revealed a new function for G3BP1 in the axon in both physiological and injured/stressed states. Interestingly, axonal G3BP1 and TIA1 only partially colocalize. This was also true for other SG components, suggesting that these G3BP1‐positive axonal puncta are distinct from SGs (Sahoo et al., 2018, 2020). Importantly, the P‐body markers DCP1A and XRN1 were absent from these axonal G3BP1‐granules (Sahoo et al., 2018). Therefore, the role of G3BP1‐granules in axons is likely related to transcript stability and/or translatability. These reports were not the first to link G3BP1 ribonucleoprotein (RNP) granules to axonal and neurite growth. In neurons, G3BP1 is localized in RNP granules containing tau mRNA, which are enriched in axons (Atlas et al., 2004). Indeed, tau mRNA has a 3’UTR sequence that targets it to axons in a microtubule‐dependent manner via RNP granules containing HuD, IMP‐1, and G3BP1 (Atlas et al., 2004). G3BP1 RNA granule formation leads to an increase in tau mRNA High Molecular Weight encoding isoforms, compared to the Low Molecular Weight encoding forms (Moschner et al., 2014). This shift in the ratio between the isoforms is associated with neurites. Finally, G3BP1 is associated in granules with the RBPs NONO, Hermes, hnRNP K, and PABP in differentiated murine retinal ganglion cell lines (RGC–5 cells) (Furukawa et al., 2015). It is suggested that these granules are transported to the neurites in association with the kinesin KIF5. However, the function of these granules is yet to be explored.
5.4.2. G3BP1 is involved in dendritic maturation
Proper spine maturation is dependent on local dendritic protein synthesis and is essential for synapses (Lai & Ip, 2003, 2009, 2013; McKinney, 2010; Yuste, 2011). mRNA localization and translational regulation requires specific RBPs, and their deficiency in neurons often leads to a more immature dendrite phenotype (more filopodia‐like spines, less mushroom‐shaped) (Afroz et al., 2017; Goetze et al., 2006; Lai & Ip, 2003, 2009, 2013). For example, in fragile X syndrome, which is caused by a deficiency of the RBP FMR1, spine morphology is noted to be more immature (Dictenberg et al., 2008; Muddashetty et al., 2011). This phenotype is mimicked by the depletion of G3BP1 whose function in spine maturation depends on PRMT8‐mediated methylation (Lo et al., 2020). Importantly, G3BP1 can rescue PRMT8‐depletion effects on dentrite morphology. These data suggest that G3BP1 impairment may induce neurodevelopmental, memory or synaptic dysfunctions in a PRMT8‐mediated manner.
6. A tale of two solitudes: G3BP1 vs. G3BP2
Since the first report, G3BP1 and its paralog G3BP2 have often been studied as one entity. Indeed, studies focused on G3BP1 frequently extend or generalize conclusions to apply to G3BP2 in the absence of evidence. In general, literature surrounding stress granule assembly and dynamics focuses on G3BP1, whereas both G3BP1 and G3BP2 has been largely implicated in cancer (Alam & Kennedy, 2019). Additionally, many studies on viral infection suggest that viruses preferentially target G3BP1 (Zhang et al., 2019). This set of work normally discounts the fact that despite the high homology among the proteins, close inspection of their protein structure suggests that G3BP protein functions may not fully overlap.
G3BP1 and G3BP2 are not isoforms of the same gene, but paralogs located on separate chromosomes: G3BP1 is localized on chromosome 5, whereas G3BP2 is situated on chromosome 4. G3BP2 can be transcribed into 19 transcripts, three of which code for proteins of 482 and 449 amino acids: the isoforms G3BP2a and G3BP2b, respectively. DNA sequence analysis reveals that G3BP2a and G3BP2b coding sequences have 60% and 55% similarity with G3BP1 (MView). At the protein level, these similarities translate to 61% and 55% identity to G3BP1 for G3BP2a and G3BP2b, respectively (MView). The most conserved region is the NTF2‐like domain, suggesting that G3BP1 and G3BP2a/b shuttle through nuclear pores and the regulation of this function is similar (Vognsen et al., 2013). Also, since the NTF2‐like domains can dimerize and are highly similar between the proteins, one could speculate that G3BP1, G3BP2a, and G3BP2b might assemble with each other to function as heterodimers or even influence homodimer functions, meaning that dysfunction in any one of the proteins may have an impact on the other. The main dissimilitude between the G3BP proteins is found in their central unstructured regions where the number of proline‐rich motifs (PxxP) varies. Specifically, G3BP2a contains a cluster of four PxxP motifs between the acid‐rich and RRM domains, whereas G3BP2b contains five in the homologous region (Kennedy et al., 2002) (Figure 1). The PxxP motifs represent the minimal SH3 domain‐binding consensus sequences (Kami et al., 2002; Saksela & Permi, 2012). Since SH3 domain‐containing proteins are pivotal signal transducers, the variability in the PxxP regions suggests that the G3BP proteins may associate with different partners to produce distinct cellular outcomes (Kurochkina & Guha, 2013; Mayer & Baltimore, 1993; Morton & Campbell, 1994; Saksela & Permi, 2012; Teyra et al., 2017). Moreover, the size of the glycine‐rich, RRM, and RGG domains varies between G3BP1 and G3BP2 (Uniprot). While in G3BP1 the glycine‐rich, RRM, and RGG domains are, respectively, of 82, 76, and 32 amino acids, in G3BP2a/b these domains are 90, 79, and 61 amino acids. Also, even though the NTF2‐like domains of G3BP1 and G3BP2 are highly similar, structural studies indicate that each have unique secondary structure (Kristensen, 2015; Vognsen et al., 2013) (Figure 1). Collectively, these structural differences suggest that G3BP1, G3BP2a, and G3BP2b are similar but sufficiently different to possibly effect distinct functions.
As paralogs, alignment and construction of a phylogenic tree across 14 species indicates that G3bp1 and G3bp2 form two distinct gene clusters that were separated from a common ancestor gene (Dereeper et al., 2008) (Figure 3). Specifically, among the various species, all the G3bp1 genes are closely related and separated from the G3bp2 genes, and vice versa. An intriguing observation is that the common ancestor gene is separated from the Drosophila and C. elegans orthologs RIN and gtbp‐1, respectively, suggesting either that some new function(s) arose during evolution or that the function(s) were separated out to two genes. Therefore, functional studies using flies and worms should likely avoid overgeneralizing to G3BP1 and G3BP2. Moreover, the evolution of G3BP1 and G3BP2 from this ancestral gene could signal that the respective proteins have acquired important new non‐redundant functions specific to high‐order species.
FIGURE 3.
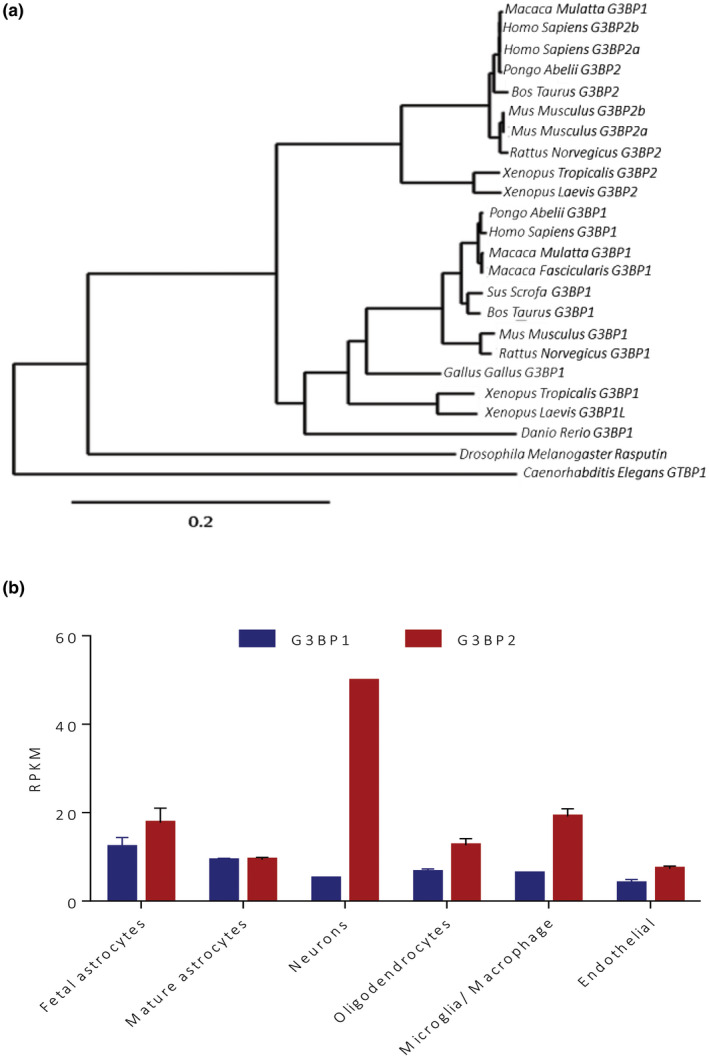
Comparisons of G3BP proteins. (a) Phylogram of 14 species comparing G3BP1 and G3BP2 orthologs. (b) Comparison of G3BP1 and G3BP2 transcriptomic levels in several brain cell subtypes, extracted from Brain RNA‐seq database in RPKM (Reads Per Kilobase of transcript per Million mapped reads) (Zhang et al., 2016).
6.1. G3BPs expression differences in the CNS
G3BP1 and G3BP2 expression levels appear to be comparable in the periphery (GTEx). However, within the CNS, G3BP2 is generally expressed at levels higher than G3BP1 (Irvine et al., 2004; Kennedy et al., 2002; Parker et al., 1996). According to public databases (GTEx, Brain RNA‐seq), depending on the brain region, G3BP2 is expressed 2‐10 times more than G3BP1. G3BP1 protein is expressed weakly throughout the brain but with the highest expression in the cerebellum (Purkinje cell layer), hippocampus (dentate granule cell layer and CA1 pyramidal regions) and the cortex (Martin et al., 2016; Parker et al., 1996). The Allen Brain Atlas also provides insight on the differential expression pattern of G3BP1 and G3BP2 (Hawrylycz et al., 2012). Specifically, while G3BP2 appears to be expressed in all layers of the brain with high expression in layers 1, 4, 5, and 6, G3BP1 is concentrated in layers 3 and 6. There also appears to be a difference among neuronal subsets such that G3BP1 is enriched in glutamatergic neurons, whereas G3BP2 is equally abundant in glutamatergic and GABAergic neurons. Inspection of transcriptomics data from several brain cell subtypes such as astrocytes, oligodendrocytes, and endothelial cells showed that these varied cell types express approximatively the same levels of G3BP1 and G3BP2 (Zhang et al., 2016) (Figure 1). The main difference between G3BP1 and G3BP2 expression lies in their levels in neurons and microglia. In both cases, G3BP2 is 10‐times more expressed than G3BP1. One could hypothesize that this differential expression may be related to specific G3BP protein functions in different cell types. Notably, exon expression analysis (GTEx) show that exon 16, the exon present in G3BP2a and not in G3BP2b, is mostly expressed in skeletal muscle, the cerebellar hemisphere, and the frontal cortex, but sparsely expressed in peripheral tissues. These differences in expression strongly suggest a difference of function for the two isoforms, with G3BP2a being the form having a potential role within the CNS.
6.2. Can G3BP2 compensate for known G3BP1 functions?
G3BP2 can induce stress granule formation similarly to G3BP1 (Matsuki et al., 2013), and complete abolishment of stress granule assembly in transformed cell lines requires genetic deletion of both G3BP1 and G3BP2 (Kedersha et al., 2016). The main difference in the structure of G3BP1 and G3BP2 is in the internal regions, which have shown to be dispensible for the formation of stress granules (Yang et al., 2020). Additionally, the protein sequence of G3BP2 has predicted phosphorylation sites that could represent S149 and S232 (Figure 1), suggesting it may also fluctuate between an open and closed conformational state, like G3BP1. However, the phase separation properties of G3BP2 are slightly different from those of G3BP1 (Guillén‐Boixet et al., 2020). In an in vitro experiment to mimic phase separation with only proteins or RNA, it was demonstrated that G3BP2 exhibits a greater tendency to phase separate with proteins and RNA compared to G3BP1, since a higher percentage of G3BP2 could form condensates with poly‐ethylene glycol (PEG) or RNA at lower concentrations (Guillén‐Boixet et al., 2020). This is probably because of G3BP2 having longer intrinsically disordered regions and RNA‐binding domain compared to G3BP1 (Figure 1). Lastly, G3BP2 is predicted to repel m6A‐modified RNA, similar to G3BP1 (Edupuganti et al., 2017). With these factors in mind, the role of G3BP2 in stress granule formation and dynamics is likely similar, if not overlapping, with G3BP1.
Given that other functions of G3BP1 are less studied for G3BP2, it is hard to predict whether G3BP2 can functionally replace G3BP1 in RQC or SMD. Although G3BP1 was the primary focus of recent papers related to RQC and SMD, there were some data in these studies to suggest that G3BP2 could have an overlapping role in these pathways as well (Anisimov et al., 2019; Fischer et al., 2020; Meyer et al., 2020). As discussed previously, G3BP1 is involved in axonal growth when aggregated in SG‐like structures and depletion of G3BP1 in the axon leads to the loss of these structures and increased axonal growth. Interestingly, G3BP2 levels were not altered in this situation, suggesting that G3BP2 cannot compensate for G3BP1 in this scenario (Sahoo et al., 2018). In the case of dendritic spine maturation, G3BP2 cannot rescue PRMT8‐depletion effects on dendrite morphology, indicating that this function is also likely unique to G3BP1 (Lo et al., 2020).
It is important to note that the RNAs bound by G3BP1 and G3BP2 are largely overlapping, although there are also many mRNA targets that are specific to only one of the G3BPs (Edupuganti et al., 2017). For example, G3BP1 uniquely binds TARDBP mRNA (which encodes TDP‐43), whereas G3BP2 uniquely binds HNRNPA1 mRNA (Edupuganti et al., 2017); both of these genes are linked to ALS and FTD. The distinctions in the mRNAs regulated by G3BP1 and G3BP2 is likely the key to defining unique functions for each protein.
6.3. G3BP2a is a regulator of IκBα/NF‐κB complexes
In neurons and glial cells, G3BP2a binds IκBα and IκBα/NF‐κB complexes leading to their cytoplasmic retention, a function that is not shared with G3BP1 (Prigent et al., 2000). G3BP2a‐mediated cytoplasmic retention of IκBα implies a positive influence on NF‐κB activation, since IκBα would be prevented from inhibiting NF‐κB in the nucleus. Conversely, G3BP2a‐mediated cytoplasmic retention of IκBα in complex with NF‐κB would imply a negative role in NF‐κB activation (Prigent et al., 2000). NF‐κB is a family of transcription factors known for their rapid and potent capacity for modulating gene expression (May & Ghosh, 1998). In mice, activation of the NF‐κB pathway in excitatory glutamatergic neurons promotes dendritic spine and excitatory synapse formation, whereas diminution of NF‐κB activity reduces dendritic spine size and density (Dresselhaus & Meffert, 2019). Thus, any dysfunction in G3BP2a could influence NF‐κB activity and ultimately be deleterious in the CNS.
NF‐κB is well known to regulate the activation of chemokines, cytokines, proinflammatory enzymes, adhesion molecules, proinflammatory transcription factors, and other factors that modulate neuronal survival (Shih et al., 2015). In addition to neurons, NF‐κB is abundant in glia where it regulates the inflammatory reaction around the neuronal environment (Aguilera et al., 2017; Engelmann & Haenold, 2016; Jha et al., 2019; Milligan & Watkins, 2009). Several neurological diseases are linked to NF‐κB activation by inflammatory mediators including ALS, FTD, and glaucoma (Swarup et al., 2011; Toth & Atkin, 2018). Loss of function mutations in the gene OPTN, which negatively regulates TNF‐α‐induced NF‐κB activation, have been reported in ALS, FTD, and glaucoma patients and up‐regulation of NF‐κB is robustly associated with AD and PD (Gveric et al., 1998; Maruyama et al., 2010; Mattson & Camandola, 2001; Minegishi et al., 2016; Terai et al., 1996; Toth & Atkin, 2018; Valerio et al., 2006). In ALS, NF‐κB becomes increasingly activated with disease progression in both SOD1 and TDP‐43 mouse models and selective inhibition of NF‐κB in microglia or neurons rescues motor neuron survival, delaying disease progression in these models (Dutta et al., 2020; Frakes et al., 2014; Haidet‐Phillips et al., 2011). Similarly, over‐expression of the NF‐κB ortholog Relish is associated with neurodegeneration in Drosophila models of AD and spinocerebellar ataxia type 3 (Li et al., 2018). As in ALS mouse models, lowering NF‐κB protein levels or activation improves phenotypes (Dutta et al., 2020; Frakes et al., 2014; Li et al., 2018).
6.4. G3BP1 and G3BP2 are Wnt pathway antagonists
Neurons and glia originate from common precursor cells (neural precursor cells, or NPCs) that proliferate in the ventricular zone of the fetal brain and spinal cord (Temple & Qian, 1996). The Wnt signaling pathway is implicated in promoting NPC self‐renewal during neural development. The over‐expression of Wnt1 and the stabilization of β‐catenin in the early stages of chick spinal cord or mouse forebrain development both induce an increased number of NPCs and suppression of neuronal differentiation (Chenn & Walsh, 2002; Megason & McMahon, 2002). The balance between proliferation and differentiation of NPCs is essential in determining the size of each region within the brain. G3BP1 is a negative regulator of Wnt/β‐catenin signaling, whereas G3BP2 is a positive regulator, giving new insights to why the two proteins are essential for development (Bikkavilli & Malbon, 2011, 2012). Moreover, impaired Wnt signaling leading to neuronal damage is observed in AD, HD, ALS and schizophrenia models (Gonzalez‐Fernandez et al., 2020; Inestrosa et al., 2005, 2012; Inestrosa & Toledo, 2008). Notably, β‐catenin levels are reduced in AD patients carrying presenilin‐1 mutations, whereas inactivation of β‐catenin is seen in HD (Godin et al., 2010; Zhang et al., 1998). Collectively, this suggests that an impairment in G3BP1 or G3BP2‐related Wnt functions has the potential to be a feature in neurological disorders, in two different ways.
7. Concluding remarks
G3BP1 has been thoroughly studied in the context of cancers and viral infections, and has now become a promising therapeutic target for specific types of tumors (Alam & Kennedy, 2019). However, the most striking observation in the G3bp1−/− mouse model was the loss of neurons, compared to any other organ (Zekri et al., 2005). This suggests that the role of G3BP1 is critical to development, and is emphasized in neurons. Therefore, we posit that the role of G3BP1 in the CNS is fundamentally different from its role in proliferating cells. Additionally, many aspects of RNA metabolism, including translation rates, decay rates, and structure, are different in neurons compared to other cell types. Importantly, we suggest that the phase separation properties of G3BP1 are impacted differently in neurons because of the higher proportion of longer transcripts, and the tissue expression differences in G3BP1. These properties of neurons, along with relatively low G3BP1 expression could be key factors in shifting the balance of G3BP1 function in stress granule assembly with its neuronal specific functions in axonal translation and dendritic spine maturation.
Finally, we submit the following points for consideration: G3BP1 and G3BP2 are structurally similar and exhibit similar LLPS behavior. Thus, it is easy to conceive that their roles in stress granule assembly and dynamics are overlapping in most contexts. However, a significant portion of the mRNAs regulated by G3BP1 and G3BP2 are different, implying that the roles of these RBPs are different in physiological conditions. Recently identified functions of G3BP1 in ribosomal recycling and structure‐mediated decay should not be generalized to G3BP2. Lastly, several important questions remain: Does G3BP2 have a role in RNA degradation? Do the transcriptomic differences between G3BP1 and G3BP2 inform on cell specific roles of each of these proteins, especially given that the function of G3BP2 is largely unexplored in the context of neurons? Are functions of G3BP1 and G3BP2, beyond their role in stress granules, impacted in neurodegenerative disease? Answering these questions will provide a number of opportunities for therapeutic interventions targeting G3BP protein function in neurodegenerative disease.
Conflict of interest
C. Vande Velde is a Handling Editor for the Journal of Neurochemistry. H. Sidibé and A. Dubinski declare no conflict of interest.
Sidibé H, Dubinski A, Vande Velde C. The multi‐functional RNA‐binding protein G3BP1 and its potential implication in neurodegenerative disease. J Neurochem. 2021;157:944–962. 10.1111/jnc.15280
Hadjara Sidibé and Alicia Dubinski contributed equally.
Funding informationResearch in the CVV laboratory is funded by CIHR, Target ALS, the ALS Canada/Brain Canada Arthur J. Hudson Translational Team Grant, and ALS Association. AD is supported by an ALS Canada Trainee Award. HS is supported by an FRQS studentship. CVV is an FRQS Senior Research Scholar.
References
- Afroz, T. , Hock, E.‐M. , Ernst, P. , Foglieni, C. , Jambeau, M. , Gilhespy, L. A. B. , Laferriere, F. , Maniecka, Z. , Plückthun, A. , Mittl, P. , Paganetti, P. , Allain, F. H. T. , & Polymenidou, M. (2017). Functional and dynamic polymerization of the ALS‐linked protein TDP‐43 antagonizes its pathologic aggregation. Nature Communications, 8(1), 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera, G. , Colín‐González, A. L. , Rangel‐López, E. , Chavarría, A. , & Santamaría, A. (2017). Redox signaling, neuroinflammation, and neurodegeneration. Antioxidants & Redox Signaling, 28(18), 1626–1651. [DOI] [PubMed] [Google Scholar]
- Alam, U. , & Kennedy, D. (2019). Rasputin a decade on and more promiscuous than ever? A review of G3BPs. Biochimica Et Biophysica Acta Molecular Cell Research, 1866(3), 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavian, K. N. , Dworetzky, S. I. , Bonanni, L. , Zhang, P. , Sacchetti, S. , Mariggio, M. A. , Onofrj, M. , Thomas, A. , Li, H. , Mangold, J. E. , Signore, A. P. , DeMarco, U. , Demady, D. R. , Nabili, P. , Lazrove, E. , Smith, P. J. S. , Gribkoff, V. K. , & Jonas, E. A. (2012). Effects of dexpramipexole on brain mitochondrial conductances and cellular bioenergetic efficiency. Brain Research, 1446, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis, A. (2006). Misregulation of tau alternative splicing in neurodegeneration and dementia. Progress in Molecular and Subcellular Biology, 44, 89–107. [DOI] [PubMed] [Google Scholar]
- Anisimov, S. , Takahashi, M. , Kakihana, T. , Katsuragi, Y. , Kitaura, H. , Zhang, L. U. , Kakita, A. , & Fujii, M. (2019). G3BP1 inhibits ubiquitinated protein aggregations induced by p62 and USP10. Scientific Reports, 9(1), 12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annibaldi, A. , Dousse, A. , Martin, S. , Tazi, J. , & Widmann, C. (2011). Revisiting G3BP1 as a RasGAP binding protein: sensitization of tumor cells to chemotherapy by the RasGAP 317–326 sequence does not involve G3BP1. PLoS One, 6(12), e29024‐e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava, Y. , Wang, Y. , Storey, J. D. , Liu, C. L. , Brown, P. O. , & Herschlag, D. (2003). Genome‐wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America, 100(7), 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas, R. , Behar, L. , Elliott, E. , & Ginzburg, I. (2004). The insulin‐like growth factor mRNA binding‐protein IMP‐1 and the Ras‐regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. Journal of Neurochemistry, 89(3), 613–626. [DOI] [PubMed] [Google Scholar]
- Aulas, A. , Caron, G. , Gkogkas, C. G. , Mohamed, N.‐V. , Destroismaisons, L. , Sonenberg, N. , Leclerc, N. , Parker, J. A. , & Vande Velde, C. (2015). G3BP1 promotes stress‐induced RNA granule interactions to preserve polyadenylated mRNA. Journal of Cell Biology, 209(1), 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas, A. , Fay, M. M. , Lyons, S. M. , Achorn, C. A. , Kedersha, N. , Anderson, P. , & Ivanov, P. (2017). Stress‐specific differences in assembly and composition of stress granules and related foci. Journal of Cell Science, 130(5), 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas, A. , Stabile, S. , & Vande Velde, C. (2012). Endogenous TDP‐43, but not FUS, contributes to stress granule assembly via G3BP. MolNeurodegener, 7, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas, A. , & Vande Velde, C. (2015). Alterations in stress granule dynamics driven by TDP‐43 and FUS: a link to pathological inclusions in ALS? Frontiers in Cellular Neuroscience, 9, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkar, N. , Kovalik, T. , Lorenzini, I. , Spangler, S. , Lacoste, A. , Sponaugle, K. , Ferrante, P. , Argentinis, E. , Sattler, R. , & Bowser, R. (2018). Artificial intelligence in neurodegenerative disease research: use of IBM Watson to identify additional RNA‐binding proteins altered in amyotrophic lateral sclerosis. Acta Neuropathologica, 135(2), 227–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmada, S. J. , Ju, S. , Arjun, A. , Batarse, A. , Archbold, H. C. , Peisach, D. , Li, X. , Zhang, Y. , Tank, E. M. H. , Qiu, H. , Huang, E. J. , Ringe, D. , Petsko, G. A. , & Finkbeiner, S. (2015). Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proceedings of the National Academy of Sciences of the United States of America, 112(25), 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, D. M. , Kaushansky, L. J. , Ward, C. L. , Sama, R. R. K. , Chian, R.‐J. , Boggio, K. J. , Quaresma, A. J. C. , Nickerson, J. A. , & Bosco, D. A. (2013). Amyotrophic lateral sclerosis‐linked FUS/TLS alters stress granule assembly and dynamics. Molecular Neurodegeneration, 8(1), 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat, V. , & Disney, M. D. (2015). RNA structures as mediators of neurological diseases and as drug targets. Neuron, 87(1), 28–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny, I. (2009). Calcium signaling and neurodegenerative diseases. Trends in Molecular Medicine, 15(3), 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikkavilli, R. K. , & Malbon, C. C. (2011). Arginine methylation of G3BP1 in response to Wnt3a regulates β‐catenin mRNA. Journal of Cell Science, 124(Pt 13), 2310–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikkavilli, R. K. , & Malbon, C. C. (2012). Wnt3a‐stimulated LRP6 phosphorylation is dependent upon arginine methylation of G3BP2. Journal of Cell Science, 125(Pt 10), 2446–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobkova, N. V. , Lyabin, D. N. , Medvinskaya, N. I. , Samokhin, A. N. , Nekrasov, P. V. , Nesterova, I. V. , Aleksandrova, I. Y. , Tatarnikova, O. G. , Bobylev, A. G. , Vikhlyantsev, I. M. , Kukharsky, M. S. , Ustyugov, A. A. , Polyakov, D. N. , Eliseeva, I. A. , Kretov, D. A. , Guryanov, S. G. , & Ovchinnikov, L. P. (2015). The Y‐box binding protein 1 suppresses Alzheimer’s disease progression in two animal models. PLoS One, 10(9), e0138867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker, G. W. , Gout, I. , Downing, A. K. , Driscoll, P. C. , Boyd, J. , Waterfield, M. D. , Campbell, I. D. (1993). Solution structure and ligand‐binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3‐kinase. Cell, 73(4), 813–822. [DOI] [PubMed] [Google Scholar]
- Bosco, D. A. , Lemay, N. , Ko, H. K. , Zhou, H. , Burke, C. , Kwiatkowski, T. J. , Sapp, P. , McKenna‐Yasek, D. , Brown, R. H. , & Hayward, L. J. (2010). Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Human Molecular Genetics, 19(21), 4160–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah, O. , Desforges, B. , Wu, T.‐D. , Pioche‐Durieu, C. , Marco, S. , Hamon, L. , Curmi, P. A. , Guerquin‐Kern, J.‐L. , Piétrement, O. , & Pastré, D. (2014). Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Research, 42(13), 8678–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. A. L. , Roberts, T. L. , Richards, R. , Woods, R. , Birrell, G. , Lim, Y. C. , Ohno, S. , Yamashita, A. , Abraham, R. T. , Gueven, N. , & Lavin, M. F. (2011). A novel role for hSMG‐1 in stress granule formation. Molecular and Cellular Biology, 31(22), 4417–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, P. , Coughlin, M. , & Mitchison, T. J. (2009). Interaction between Poly(ADP‐ribose) and NuMA contributes to mitotic spindle pole assembly. Molecular Biology of the Cell, 20(21), 4575–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Dumelie, J. G. , Li, X. , Cheng, M. H. K. , Yang, Z. , Laver, J. D. , Siddiqui, N. U. , Westwood, J. T. , Morris, Q. , Lipshitz, H. D. , & Smibert, C. A. (2014). Global regulation of mRNA translation and stability in the early Drosophilaembryo by the Smaug RNA‐binding protein. Genome Biology, 15(1), R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn, A. , & Walsh, C. A. (2002). Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science, 297(5580), 365–369. [DOI] [PubMed] [Google Scholar]
- Chung, J.‐W. , Ryu, W.‐S. , Kim, B. J. , & Yoon, B.‐W. (2015). Elevated calcium after acute ischemic stroke: association with a poor short‐term outcome and long‐term mortality. Journal of Stroke, 17(1), 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléry, A. , Blatter, M. , & Allain, F. H. (2008). RNA recognition motifs: boring? Not Quite. Current Opinion in Structural Biology, 18(3), 290–298. [DOI] [PubMed] [Google Scholar]
- Conrad, C. , Zhu, J. , Conrad, C. , Schoenfeld, D. , Fang, Z. , Ingelsson, M. , Stamm, S. , Church, G. , & Hyman, B. T. (2007). Single molecule profiling of tau gene expression in Alzheimer's disease. Journal of Neurochemistry, 103(3), 1228–1236. [DOI] [PubMed] [Google Scholar]
- Dereeper, A. , Guignon, V. , Blanc, G. , Audic, S. , Buffet, S. , Chevenet, F. , Dufayard, J.‐F. , Guindon, S. , Lefort, V. , Lescot, M. , Claverie, J.‐M. , & Gascuel, O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non‐specialist. Nucleic Acids Research, 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg, J. B. , Swanger, S. A. , Antar, L. N. , Singer, R. H. , & Bassell, G. J. (2008). A direct role for FMRP in activity‐dependent dendritic mRNA transport links filopodial‐spine morphogenesis to fragile X syndrome. Developmental Cell, 14(6), 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, N. , Chen, J. , Yu, S. , Gao, Y. , & Li, Y. (2016). G3BP1 contributes to tumor metastasis via upregulation of Slug expression in hepatocellular carcinoma. American Journal of Cancer Research, 6(11), 2641–2650. [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus, E. C. , & Meffert, M. K. (2019). Cellular specificity of NF‐κB function in the nervous system. Frontiers in Immunology, 10, 1043‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y. , Du, A. , Gu, J. , Duan, G. , Wang, C. , Gui, X. , Ma, Z. , Qian, B. , Deng, X. , Zhang, K. , Sun, L. E. , Tian, K. , Zhang, Y. , Jiang, H. , Liu, C. , & Fang, Y. (2019). PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease‐related RNA‐binding proteins. Cell Research, 29(3), 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, K. , Thammisetty, S. S. , Boutej, H. , Bareil, C. , & Julien, J.‐P. (2020). Mitigation of ALS pathology by neuron‐specific inhibition of nuclear factor kappa B signaling. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience, 40(26), 5137–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti, R. R. , Geiger, S. , Lindeboom, R. G. H. , Shi, H. , Hsu, P. J. , Lu, Z. , Wang, S.‐Y. , Baltissen, M. P. A. , Jansen, P. W. T. C. , Rossa, M. , Müller, M. , Stunnenberg, H. G. , He, C. , Carell, T. , & Vermeulen, M. (2017). N6‐methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nature Structural & Molecular Biology, 24, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann, C. , & Haenold, R. (2016). Transcriptional control of synaptic plasticity by transcription factor NF‐κB. Neural Plasticity, 2016, 7027949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér, Á. , Juhász, A. , László, A. , Kálmán, J. , Pákáski, M. , Kálmán, J. , & Janka, Z. (2012). Association between a variant of the sigma‐1 receptor gene and Alzheimer's disease. Neuroscience Letters, 517(2), 136–139. [DOI] [PubMed] [Google Scholar]
- Fiesel, F. C. , Voigt, A. , Weber, S. S. , Van den Haute, C. , Waldenmaier, A. , Görner, K. , Walter, M. , Anderson, M. L. , Kern, J. V. , Rasse, T. M. , Schmidt, T. , Springer, W. , Kirchner, R. , Bonin, M. , Neumann, M. , Baekelandt, V. , Alunni‐Fabbroni, M. , Schulz, J. B. , & Kahle, P. J. (2010). Knockdown of transactive response DNA‐binding protein (TDP‐43) downregulates histone deacetylase 6. The EMBO Journal, 29(1), 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, J. W. , Busa, V. F. , Shao, Y. , & Leung, A. K. L. (2020). Structure‐mediated RNA decay by UPF1 and G3BP1. Molecular Cell, 78(1), 70–84.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman, M. S. , Trojanowski, J. Q. , & Lee, V. M. Y. (2007). TDP‐43: a novel neurodegenerative proteinopathy. Current Opinion in Neurobiology, 17(5), 548–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frakes, A. E. , Ferraiuolo, L. , Haidet‐Phillips, A. M. , Schmelzer, L. , Braun, L. , Miranda, C. J. , Ladner, K. J. , Bevan, A. K. , Foust, K. D. , Godbout, J. P. , Popovich, P. G. , Guttridge, D. C. , & Kaspar, B. K. (2014). Microglia induce motor neuron death via the classical NF‐κB pathway in amyotrophic lateral sclerosis. Neuron, 81(5), 1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. , & Zhuang, X. (2020). m(6)A‐binding YTHDF proteins promote stress granule formation. Nature Chemical Biology, 16(9), 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, M. T. , Sakamoto, H. , & Inoue, K. (2015). Interaction and colocalization of HERMES/RBPMS with NonO, PSF, and G3BP1 in neuronal cytoplasmic RNP granules in mouse retinal line cells. Genes to Cells, 20(4), 257–266. [DOI] [PubMed] [Google Scholar]
- Gal, J. , Chen, J. , Na, D. Y. , Tichacek, L. , Barnett, K. R. , & Zhu, H. (2019). The acetylation of lysine‐376 of G3BP1 regulates RNA binding and stress granule dynamics. Molecular and Cellular Biology, 39(22), e00052‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal, J. , Zhang, J. , Kwinter, D. M. , Zhai, J. , Jia, H. , Jia, J. , & Zhu, H. (2010). Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. NeurobiolAging, 32(12), 2323.e27‐.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, A. , Lozano, G. , Piñeiro, D. , & Martinez‐Salas, E. (2017). G3BP1 interacts directly with the FMDV IRES and negatively regulates translation. The FEBS Journal, 284(19), 3202–3217. [DOI] [PubMed] [Google Scholar]
- Gallo, J. M. , Noble, W. , & Martin, T. R. (2007). RNA and protein‐dependent mechanisms in tauopathies: consequences for therapeutic strategies. Cellular and Molecular Life Sciences, 64(13), 1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz, D. C. , Rujescu, D. , Tang, Y. , Berendt, F. J. , Hartmann, A. M. , Faltraco, F. , Rosenberg, C. , Hulette, C. , Jellinger, K. , Hampel, H. , Riederer, P. , Moller, H.‐J. , Andreadis, A. , Henkel, K. , & Stamm, S. (2006). The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2‐BETA1 changes in sporadic Alzheimer's disease. Journal of Neurochemistry, 96(3), 635–644. [DOI] [PubMed] [Google Scholar]
- Godin, J. D. , Poizat, G. , Hickey, M. A. , Maschat, F. , & Humbert, S. (2010). Mutant huntingtin‐impaired degradation of beta‐catenin causes neurotoxicity in Huntington's disease. The EMBO Journal, 29(14), 2433–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze, B. , Tuebing, F. , Xie, Y. , Dorostkar, M. M. , Thomas, S. , Pehl, U. , Boehm, S. , Macchi, P. , & Kiebler, M. A. (2006). The brain‐specific double‐stranded RNA‐binding protein Staufen2 is required for dendritic spine morphogenesis. The Journal of Cell Biology, 172(2), 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Fernandez, C. , González, P. , & Rodríguez, F. J. (2020). New insights into Wnt signaling alterations in amyotrophic lateral sclerosis: a potential therapeutic target? Neural Regeneration Research, 15(9), 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregianin, E. , Pallafacchina, G. , Zanin, S. , Crippa, V. , Rusmini, P. , Poletti, A. , Fang, M. , Li, Z. , Diano, L. , Petrucci, A. , & Lispi, L. (2016). Loss‐of‐function mutations in the SIGMAR1 gene cause distal hereditary motor neuropathy by impairing ER‐mitochondria tethering and Ca2+ signalling. Human Molecular Genetics, 25(17), 3741–3753. [DOI] [PubMed] [Google Scholar]
- Guillén‐Boixet, J. , Kopach, A. , Holehouse, A. S. , Wittmann, S. , Jahnel, M. , Schlüßler, R. , Kim, K. , Trussina, I. R. E. A. , Wang, J. , Mateju, D. , Poser, I. , Maharana, S. , Ruer‐Gruß, M. , Richter, D. , Zhang, X. , Chang, Y.‐T. , Guck, J. , Honigmann, A. , Mahamid, J. , Hyman, A. A. , Pappu, R. V. , Alberti, S. , & Franzmann, T. M. (2020). RNA‐induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell, 181(2), 346–61.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, A. , Gu, H. , Zhou, J. , Mulhern, D. , Wang, Y. I. , Lee, K. A. , Yang, V. , Aguiar, M. , Kornhauser, J. , Jia, X. , Ren, J. , Beausoleil, S. A. , Silva, J. C. , Vemulapalli, V. , Bedford, M. T. , & Comb, M. J. (2014). Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Molecular & Cellular Proteomics, 13(1), 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gveric, D. , Kaltschmidt, C. , Cuzner, M. L. , & Newcombe, J. (1998). Transcription factor NF‐kappaB and inhibitor I kappaBalpha are localized in macrophages in active multiple sclerosis lesions. Journal of Neuropathology and Experimental Neurology, 57(2), 168–178. [DOI] [PubMed] [Google Scholar]
- Haidet‐Phillips, A. M. , Hester, M. E. , Miranda, C. J. , Meyer, K. , Braun, L. , Frakes, A. , Song, S. W. , Likhite, S. , Murtha, M. J. , Foust, K. D. , Rao, M. , Eagle, A. , Kammesheidt, A. , Christensen, A. , Mendell, J. R. , Burghes, A. H. M. , & Kaspar, B. K. (2011). Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. NatBiotechnol, 29(9), 824–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz, M. J. , Lein, E. S. , Guillozet‐Bongaarts, A. L. , Shen, E. H. , Ng, L. , Miller, J. A. , van de Lagemaat, L. N. , Smith, K. A. , Ebbert, A. , Riley, Z. L. , Abajian, C. , Beckmann, C. F. , Bernard, A. , Bertagnolli, D. , Boe, A. F. , Cartagena, P. M. , Chakravarty, M. M. , Chapin, M. , Chong, J. , … Jones, A. R. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 489(7416), 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inestrosa, N. C. , Godoy, J. A. , Quintanilla, R. A. , Koenig, C. S. , & Bronfman, M. (2005). Peroxisome proliferator‐activated receptor γ is expressed in hippocampal neurons and its activation prevents β‐amyloid neurodegeneration: role of Wnt signaling. Experimental Cell Research, 304(1), 91–104. [DOI] [PubMed] [Google Scholar]
- Inestrosa, N. C. , Montecinos‐Oliva, C. , & Fuenzalida, M. (2012). Wnt signaling: Role in Alzheimer disease and schizophrenia. Journal of Neuroimmune Pharmacology, 7(4), 788–807. [DOI] [PubMed] [Google Scholar]
- Inestrosa, N. C. , & Toledo, E. M. (2008). The role of Wnt signaling in neuronal dysfunction in Alzheimer's disease. Molecular Neurodegeneration, 3(1), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine, K. , Stirling, R. , Hume, D. , & Kennedy, D. (2004). Rasputin, more promiscuous than ever: a review of G3BP. International Journal of Developmental Biology, 48(10), 1065–1077. [DOI] [PubMed] [Google Scholar]
- Isabelle, M. , Gagné, J.‐P. , Gallouzi, I.‐E. , & Poirier, G. G. (2012). Quantitative proteomics and dynamic imaging reveal that G3BP‐mediated stress granule assembly is poly(ADP‐ribose)‐dependent following exposure to MNNG‐induced DNA alkylation. Journal of Cell Science, 125(19), 4555. [DOI] [PubMed] [Google Scholar]
- Jain, S. , Wheeler, J. R. , Walters, R. W. , Agrawal, A. , Barsic, A. , & Parker, R. (2016). ATPase‐modulated stress granules contain a diverse proteome and substructure. Cell, 164(3), 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je, G. , Guhathakurta, S. , Yun, S. P. , Ko, H. S. , & Kim, Y.‐S. (2018). A novel extended form of alpha‐synuclein 3′UTR in the human brain. Molecular Brain, 11(1), 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, N. K. , Jha, S. K. , Kar, R. , Nand, P. , Swati, K. , & Goswami, V. K. (2019). Nuclear factor‐kappa β as a therapeutic target for Alzheimer's disease. Journal of Neurochemistry, 150(2), 113–137. [DOI] [PubMed] [Google Scholar]
- Joazeiro, C. A. P. (2019). Mechanisms and functions of ribosome‐associated protein quality control. Nature Reviews Molecular Cell Biology, 20(6), 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami, K. , Takeya, R. , Sumimoto, H. , & Kohda, D. (2002). Diverse recognition of non‐PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p. The EMBO Journal, 21(16), 4268–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, A. , Fushimi, K. , Zhou, X. , Ray, P. , Shi, C. , Chen, X. , Liu, Z. , Chen, S. , & Wu, J. Y. (2011). RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem‐loop structure at the 5' splice site. Molecular and Cellular Biology, 31(9), 1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto, E. M. , Vivar, C. , & Camandola, S. (2012). Physiology and pathology of calcium signaling in the brain. Frontiers in Pharmacology, 3, 61‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. , Panas, M. D. , Achorn, C. A. , Lyons, S. , Tisdale, S. , Hickman, T. , Thomas, M. , Lieberman, J. , McInerney, G. M. , Ivanov, P. , & Anderson, P. (2016). G3BP‐Caprin1‐USP10 complexes mediate stress granule condensation and associate with 40S subunits. The Journal of Cell Biology, 212(7), 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, D. , French, J. , Guitard, E. , Ru, K. , Tocque, B. , & Mattick, J. (2002). Characterization of G3BPs: Tissue specific expression, chromosomal localisation and rasGAP120 binding studies. Journal of Cellular Biochemistry, 84(1), 173–187. [DOI] [PubMed] [Google Scholar]
- Kertesz, A. (2010). Frontotemporal dementia. Pick's Disease. Ideggyogysz, 63(1–2), 4–12. [PubMed] [Google Scholar]
- Khong, A. , Kerr, C. H. , Yeung, C. H. L. , Keatings, K. , Nayak, A. , Allan, D. W. , & Jan, E. (2017). Disruption of stress granule formation by the multifunctional cricket paralysis virus 1A protein. Journal of Virology, 91(5), e01779‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong, A. , & Parker, R. (2018). mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. The Journal of Cell Biology, 217, 4124–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klim, J. R. , Williams, L. A. , Limone, F. , Guerra San Juan, I. , Davis‐Dusenbery, B. N. , Mordes, D. A. , Burberry, A. , Steinbaugh, M. J. , Gamage, K. K. , Kirchner, R. , Moccia, R. , Cassel, S. H. , Chen, K. , Wainger, B. J. , Woolf, C. J. , & Eggan, K. (2019). ALS‐implicated protein TDP‐43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nature Neuroscience, 22(2), 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotova, E. , Jarnik, M. , & Tulin, A. V. (2009). Poly (ADP‐ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genetics, 5(2), e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousaka, A. , Mori, Y. , Koyama, Y. , Taneda, T. , Miyata, S. , & Tohyama, M. (2009). The distribution and characterization of endogenous protein arginine N‐methyltransferase 8 in mouse CNS. Neuroscience, 163(4), 1146–1157. [DOI] [PubMed] [Google Scholar]
- Kristensen, O. (2015). Crystal structure of the G3BP2 NTF2‐like domain in complex with a canonical FGDF motif peptide. Biochemical and Biophysical Research Communications, 467(1), 53–57. [DOI] [PubMed] [Google Scholar]
- Kurochkina, N. , & Guha, U. (2013). SH3 domains: modules of protein‐protein interactions. Biophys Rev, 5(1), 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S. , Zhang, Y. , & Matthias, P. (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes & Development, 21(24), 3381–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier‐Tourenne, C. , Polymenidou, M. , Hutt, K. R. , Vu, A. Q. , Baughn, M. , Huelga, S. C. , Clutario, K. M. , Ling, S.‐C. , Liang, T. Y. , Mazur, C. , Wancewicz, E. , Kim, A. S. , Watt, A. , Freier, S. , Hicks, G. G. , Donohue, J. P. , Shiue, L. , Bennett, C. F. , Ravits, J. , Cleveland, D. W. , & Yeo, G. W. (2012). Divergent roles of ALS‐linked proteins FUS/TLS and TDP‐43 intersect in processing long pre‐mRNAs. Nature Neuroscience, 15(11), 1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, K. O. , & Ip, N. Y. (2003). Central synapse and neuromuscular junction: same players, different roles. Trends in Genetics, 19(7), 395–402. [DOI] [PubMed] [Google Scholar]
- Lai, K. O. , & Ip, N. Y. (2009). Synapse development and plasticity: roles of ephrin/Eph receptor signaling. Current Opinion in Neurobiology, 19(3), 275–283. [DOI] [PubMed] [Google Scholar]
- Lai, K.‐O. , & Ip, N. Y. (2013). Structural plasticity of dendritic spines: The underlying mechanisms and its dysregulation in brain disorders. Biochimica Et Biophysica Acta (BBA) ‐ Molecular Basis of Disease, 1832(12), 2257–2263. [DOI] [PubMed] [Google Scholar]
- Laver, J. D. , Ly, J. , Winn, A. K. , Karaiskakis, A. , Lin, S. , Nie, K. , Benic, G. , Jaberi‐Lashkari, N. , Cao, W. X. , Khademi, A. , Westwood, J. T. , Sidhu, S. S. , Morris, Q. , Angers, S. , Smibert, C. A. , & Lipshitz, H. D. (2020). The RNA‐binding protein rasputin/G3BP enhances the stability and translation of its target mRNAs. Cell Reports, 30(10), 3353–67.e7. [DOI] [PubMed] [Google Scholar]
- Lee, A. K. , Klein, J. , Fon Tacer, K. , Lord, T. , Oatley, M. J. , Oatley, J. M. , Porter, S. N. , Pruett‐Miller, S. M. , Tikhonova, E. B. , Karamyshev, A. L. , Wang, Y.‐D. , Yang, P. , Korff, A. , Kim, H. J. , Taylor, J. P. , & Potts, P. R. (2020). Translational repression of G3BP in cancer and germ cells suppresses stress granules and enhances stress tolerance. Molecular Cell, 79(4), 645–59.e9. [DOI] [PubMed] [Google Scholar]
- Leung, A. K. L. , Vyas, S. , Rood, J. E. , Bhutkar, A. , Sharp, P. A. , & Chang, P. (2011). Poly(ADP‐ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular Cell, 42(4), 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. R., King O. D., Shorter J., & Gitler A. D. (2013) Stress granules as crucibles of ALS pathogenesis. Journal of Cell Biology 201(3), 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. X. , Sibon, O. C. M. , & Dijkers, P. F. (2018). Inhibition of NF‐κB in astrocytes is sufficient to delay neurodegeneration induced by proteotoxicity in neurons. Journal of Neuroinflammation, 15(1), 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R. E. (2016). Enterovirus control of translation and RNA granule stress responses. Viruses, 8(4), 93‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, L.‐H.‐Y. , Dong, R. , Lyu, Q. , & Lai, K.‐O. (2020). The Protein arginine methyltransferase PRMT8 and substrate G3BP1 Control Rac1‐PAK1 signaling and actin cytoskeleton for dendritic spine maturation. Cell Reports, 31(10), 107744. [DOI] [PubMed] [Google Scholar]
- Luty, A. A. , Kwok, J. B. J. , Dobson‐Stone, C. , Loy, C. T. , Coupland, K. G. , Karlström, H. , Sobow, T. , Tchorzewska, J. , Maruszak, A. , Barcikowska, M. , Panegyres, P. K. , Zekanowski, C. , Brooks, W. S. , Williams, K. L. , Blair, I. P. , Mather, K. A. , Sachdev, P. S. , Halliday, G. M. , & Schofield, P. R. (2010). Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration‐motor neuron disease. Annals of Neurology, 68(5), 639–649. [DOI] [PubMed] [Google Scholar]
- Makeyev, E. V. , Zhang, J. , Carrasco, M. A. , & Maniatis, T. (2007). The MicroRNA miR‐124 promotes neuronal differentiation by triggering brain‐specific alternative pre‐mRNA splicing. Molecular Cell, 27(3), 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller, S. , Soltanieh, S. , Server, K. L. , Mak, R. , Jin, W. , Fang, M. Y. , Luo, E.‐C. , Krach, F. , Yang, D. , Sen, A. , Fulzele, A. , Wozniak, J. M. , Gonzalez, D. J. , Kankel, M. W. , Gao, F.‐B. , Bennett, E. J. , Lécuyer, E. , & Yeo, G. W. (2018). Context‐dependent and disease‐specific diversity in protein interactions within stress granules. Cell, 172(3), 590–604.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. , Bellora, N. , González‐Vallinas, J. , Irimia, M. , Chebli, K. , de Toledo, M. , Raabe, M. , Eyras, E. , Urlaub, H. , Blencowe, B. J. , & Tazi, J. (2016). Preferential binding of a stable G3BP ribonucleoprotein complex to intron‐retaining transcripts in mouse brain and modulation of their expression in the cerebellum. Journal of Neurochemistry, 139(3), 349–368. [DOI] [PubMed] [Google Scholar]
- Martin, S. , Zekri, L. , Metz, A. , Maurice, T. , Chebli, K. , Vignes, M. , & Tazi, J. (2013). Deficiency of G3BP1, the stress granules assembly factor, results in abnormal synaptic plasticity and calcium homeostasis in neurons. Journal of Neurochemistry, 125(2), 175–184. [DOI] [PubMed] [Google Scholar]
- Maruszak, A. , Safranow, K. , Gacia, M. , Gabryelewicz, T. , Słowik, A. , Styczyńska, M. , Pepłońska, B. , Golan, M. P. , Żekanowski, C. , & Barcikowska, M. (2007). Sigma receptor type 1 gene variation in a group of Polish patients with Alzheimer's disease and mild cognitive impairment. Dementia and Geriatric Cognitive Disorders, 23(6), 432–438. [DOI] [PubMed] [Google Scholar]
- Maruyama, H. , Morino, H. , Ito, H. , Izumi, Y. , Kato, H. , Watanabe, Y. , Kinoshita, Y. , Kamada, M. , Nodera, H. , Suzuki, H. , Komure, O. , Matsuura, S. , Kobatake, K. , Morimoto, N. , Abe, K. , Suzuki, N. , Aoki, M. , Kawata, A. , Hirai, T. , … Kawakami, H. (2010). Mutations of optineurin in amyotrophic lateral sclerosis. Nature, 465(7295), 223–226. [DOI] [PubMed] [Google Scholar]
- Matheny, T. , Van Treeck, B. , Huynh, T. N. , & Parker, R. (2020). RNA partitioning into stress granules is based on the summation of multiple interactions. bioRxiv, 2020(04), pp. 15.043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki, H. , Takahashi, M. , Higuchi, M. , Makokha, G. N. , Oie, M. , & Fujii, M. (2013). Both G3BP1 and G3BP2 contribute to stress granule formation. Genes to Cells, 18(2), 135–146. [DOI] [PubMed] [Google Scholar]
- Mattson, M. P. (2007). Calcium and neurodegeneration. Aging Cell, 6(3), 337–350. [DOI] [PubMed] [Google Scholar]
- Mattson, M. P. , & Camandola, S. (2001). NF‐κB in neuronal plasticity and neurodegenerative disorders. The Journal of Clinical Investigation, 107(3), 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, M. J. , & Ghosh, S. (1998). Signal transduction through NF‐kappa B. Immunology Today, 19(2), 80–88. [DOI] [PubMed] [Google Scholar]
- Mayer, B. J. , & Baltimore, D. (1993). Signalling through SH2 and SH3 domains. Trends in Cell Biology, 3(1), 8–13. [DOI] [PubMed] [Google Scholar]
- McBride, A. E. , & Silver, P. A. (2001). State of the arg: protein methylation at arginine comes of age. Cell, 106(1), 5–8. [DOI] [PubMed] [Google Scholar]
- McDonald, K. K. , Aulas, A. , Destroismaisons, L. , Pickles, S. , Beleac, E. , Camu, W. , Rouleau, G. A. , & Vande Velde, C. (2011). TAR DNA‐binding protein 43 (TDP‐43) regulates stress granule dynamics via differential regulation of G3BP and TIA‐1. HumMolGenet, 20(7), 1400–1410. [DOI] [PubMed] [Google Scholar]
- McKinney, R. A. (2010). Excitatory amino acid involvement in dendritic spine formation, maintenance and remodelling. The Journal of Physiology, 588(1), 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason, S. G. , & McMahon, A. P. (2002). A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development, 129(9), 2087. [DOI] [PubMed] [Google Scholar]
- Memczak, S. , Jens, M. , Elefsinioti, A. , Torti, F. , Krueger, J. , Rybak, A. , Maier, L. , Mackowiak, S. D. , Gregersen, L. H. , Munschauer, M. , Loewer, A. , Ziebold, U. , Landthaler, M. , Kocks, C. , le Noble, F. , & Rajewsky, N. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), 333–338. [DOI] [PubMed] [Google Scholar]
- Meyer, C. , Garzia, A. , Morozov, P. , Molina, H. , & Tuschl, T. (2020). The G3BP1‐family‐USP10 deubiquitinase complex rescues ubiquitinated 40S subunits of ribosomes stalled in translation from lysosomal degradation. Molecular Cell, 77(6), 1193–205.e5. [DOI] [PubMed] [Google Scholar]
- Middleton, S. A. , Eberwine, J. , & Kim, J. (2019). Comprehensive catalog of dendritically localized mRNA isoforms from sub‐cellular sequencing of single mouse neurons. BMC Biology, 17(1), 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, E. D. , & Watkins, L. R. (2009). Pathological and protective roles of glia in chronic pain. Nature Reviews Neuroscience, 10(1), 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi, Y. , Nakayama, M. , Iejima, D. , Kawase, K. , & Iwata, T. (2016). Significance of optineurin mutations in glaucoma and other diseases. Progress in Retinal and Eye Research, 55, 149–181. [DOI] [PubMed] [Google Scholar]
- Mori, T. , Hayashi, T. , & Su, T. P. (2012). Compromising σ‐1 receptors at the endoplasmic reticulum render cytotoxicity to physiologically relevant concentrations of dopamine in a nuclear factor‐κB/Bcl‐2‐dependent mechanism: potential relevance to Parkinson's disease. Journal of Pharmacology and Experimental Therapeutics, 341(3), 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, C. J. , & Campbell, I. D. (1994). SH3 domains. Molecular 'velcro'. Current Biology: CB, 4(7), 615–617. [DOI] [PubMed] [Google Scholar]
- Moschner, K. , Sundermann, F. , Meyer, H. , da Graca, A. P. , Appel, N. , Paululat, A. , Bakota, L. , & Brandt, R. (2014). RNA protein granules modulate tau isoform expression and induce neuronal sprouting. The Journal of Biological Chemistry, 289(24), 16814–16825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty, R. S. , Nalavadi, V. C. , Gross, C. , Yao, X. , Xing, L. , Laur, O. , Warren, S. T. , & Bassell, G. J. (2011). Reversible inhibition of PSD‐95 mRNA translation by miR‐125a, FMRP phosphorylation, and mGluR signaling. Molecular Cell, 42(5), 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass, U. , & Blobel, G. (1996). Role of the nuclear transport factor p10 in nuclear import. Science, 272(5258), 120–122. [DOI] [PubMed] [Google Scholar]
- Parker, F. , Maurier, F. , Delumeau, I. , Duchesne, M. , Faucher, D. , Debussche, L. , Dugue, A. , Schweighoffer, F. , & Tocque, B. (1996). A Ras‐GTPase‐activating protein SH3‐domain‐binding protein. Molecular and Cellular Biology, 16(6), 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva, V. P. , Skabkin, M. A. , Hellen, C. U. T. , Pestova, T. V. , & Pisarev, A. V. (2011). Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. The EMBO Journal, 30(9), 1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka, M. , Glažar, P. , Hernandez‐Miranda, L. R. , Memczak, S. , Wolf, S. A. , Rybak‐Wolf, A. , Filipchyk, A. , Klironomos, F. , Cerda Jara, C. A. , Fenske, P. , Trimbuch, T. , Zywitza, V. , Plass, M. , Schreyer, L. , Ayoub, S. , Kocks, C. , Kühn, R. , Rosenmund, C. , Birchmeier, C. , & Rajewsky, N. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science, 357(6357), eaam8526. [DOI] [PubMed] [Google Scholar]
- Prigent, M. , Barlat, I. , Langen, H. , & Dargemont, C. (2000). IkappaBalpha and IkappaBalpha /NF‐kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3‐binding protein 2. The Journal of Biological Chemistry, 275(46), 36441–36449. [DOI] [PubMed] [Google Scholar]
- Ries, R. J. , Zaccara, S. , Klein, P. , Olarerin‐George, A. , Namkoong, S. , Pickering, B. F. , Patil, D. P. , Kwak, H. , Lee, J. H. , & Jaffrey, S. R. (2019). m6A enhances the phase separation potential of mRNA. Nature, 571(7765), 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo, P. K. , Kar, A. N. , Samra, N. , Terenzio, M. , Patel, P. , Lee, S. J. , Miller, S. , Thames, E. , Jones, B. , Kawaguchi, R. , Coppola, G. , Fainzilber, M. , & Twiss, J. L. (2020). A Ca2+‐dependent switch activates axonal casein kinase 2α translation and drives G3BP1 granule disassembly for axon regeneration. Current Biology, 30(24), 4882–4895.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo, P. K. , Lee, S. J. , Jaiswal, P. B. , Alber, S. , Kar, A. N. , Miller‐Randolph, S. , Taylor, E. E. , Smith, T. , Singh, B. , Ho, T.‐Y. , Urisman, A. , Chand, S. , Pena, E. A. , Burlingame, A. L. , Woolf, C. J. , Fainzilber, M. , English, A. W. , & Twiss, J. L. (2018). Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nature Communications, 9(1), 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksela, K. , & Permi, P. (2012). SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Letters, 586(17), 2609–2614. [DOI] [PubMed] [Google Scholar]
- Sanders, D. W. , Kedersha, N. , Lee, D. S. W. , Strom, A. R. , Drake, V. , Riback, J. A. , Bracha, D. , Eeftens, J. M. , Iwanicki, A. , Wang, A. , Wei, M.‐T. , Whitney, G. , Lyons, S. M. , Anderson, P. , Jacobs, W. M. , Ivanov, P. , & Brangwynne, C. P. (2020). Competing Protein‐RNA interaction networks control Multiphase Intracellular Organization. Cell, 181(2), 306–24.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, V. , Dantzer, F. , Ame, J. C. , & de Murcia, G. (2006). Poly(ADP‐ribose): novel functions for an old molecule. Nature Reviews Molecular Cell Biology, 7(7), 517–528. [DOI] [PubMed] [Google Scholar]
- Scotter, E. L. , Chen, H. J. , & Shaw, C. E. (2015). TDP‐43 proteinopathy and ALS: insights into disease mechanisms and therapeutic targets. Neurotherapeutics: the Journal of the American Society for Experimental NeuroTherapeutics, 12(2), 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton, C. F. , Tang, A. A. , Kulkarni, A. , West, J. , Brooks, M. , Stubblefield, J. J. , Liu, Y. , Zhang, M. Q. , Green, C. B. , Huber, K. M. , Huang, E. J. , Herz, J. , & Yu, G. (2014). Activity‐dependent FUS dysregulation disrupts synaptic homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 111(44), E4769–E4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, R.‐H. , Wang, C.‐Y. , & Yang, C.‐M. (2015). NF‐kappaB signaling pathways in neurological inflammation: A mini review. Frontiers in Molecular Neuroscience, 8, 77‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, C. J. , Eyler, D. E. , & Green, R. (2010). Dom34:Hbs1 promotes subunit dissociation and peptidyl‐tRNA drop‐off to initiate no‐go decay. Science (New York, NY), 330(6002), 369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidibé, H. , Khalfallah, Y. , Xiao, S. , Gómez, N. B. , Tank, E. M. H. , Di Tomasso, G. , Bareke, E. , Aulas, A. , McKeever, P. M. , Melamed, Z. E. , Deshaies, J.‐E. , Zinman, L. , Parker, J. A. , Legault, P. , Tétreault, M. , Barmada, S. J. , Robertson, J. , & Vande Velde, C. (2020). TDP‐43 stabilizes transcripts encoding stress granule protein G3BP1: potential relevance to ALS/FTD. bioRxiv, 2020(09), pp. 15.298455. [Google Scholar]
- Sidibé, H. , & Vande Velde, C. (2019). RNA granules and their role in neurodegenerative diseases. In Oeffinger M., & Zenklusen D. (Eds.), The Biology of mRNA: Structure and Function (pp. 195–245). Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- Simandi, Z. , Pajer, K. , Karolyi, K. , Sieler, T. , Jiang, L.‐L. , Kolostyak, Z. , Sari, Z. , Fekecs, Z. , Pap, A. , Patsalos, A. , Contreras, G. A. , Reho, B. , Papp, Z. , Guo, X. , Horvath, A. , Kiss, G. , Keresztessy, Z. , Vámosi, G. , Hickman, J. , … Nagy, L. (2018). Arginine methyltransferase PRMT8 provides cellular stress tolerance in aging motoneurons. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience, 38(35), 7683–7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões‐Pires, C. , Zwick, V. , Nurisso, A. , Schenker, E. , Carrupt, P.‐A. , & Cuendet, M. (2013). HDAC6 as a target for neurodegenerative diseases: what makes it different from the other HDACs? Molecular Neurodegeneration, 8(1), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, S. , Xu, Y. , Wang, B. , David, M. D. , Schubert, P. , Kennedy, D. & Schrader, J. W. (2007). Distinct structural features of caprin‐1 mediate its interaction with G3BP‐1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Molecular and Cellular Biology, 27(6), 2324–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan, S. P. , El‐Naggar, A. , Leprivier, G. , Cheng, H. , Hajee, S. , Grunewald, T. G. P. , Zhang, F. , Ng, T. , Delattre, O. , Evdokimova, V. , Wang, Y. , Gleave, M. , & Sorensen, P. H. (2015). YB‐1 regulates stress granule formation and tumor progression by translationally activating G3BP1. The Journal of Cell Biology, 208(7), 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan, S. P. , Zhang, F. , Saxena, N. , Huang, J. N. , Kuo, I.‐C. , Low, C. , Bell, R. , Adomat, H. , Stoynov, N. , Foster, L. , Gleave, M. , & Sorensen, P. H. (2020). G3BP1‐linked mRNA partitioning supports selective protein synthesis in response to oxidative stress. Nucleic Acids Research, 48(12), 6855–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, V. , Phaneuf, D. , Dupre, N. , Petri, S. , Strong, M. , Kriz, J. , Julien, J. P. (2011). Deregulation of TDP‐43 in amyotrophic lateral sclerosis triggers nuclear factor kappaB‐mediated pathogenic pathways. JExpMed, 208(12), 2429–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagashira, H. , Shinoda, Y. , Shioda, N. , & Fukunaga, K. (2014). Methyl pyruvate rescues mitochondrial damage caused by SIGMAR1 mutation related to amyotrophic lateral sclerosis. Biochimica Et Biophysica Acta, 1840(12), 3320–3334. [DOI] [PubMed] [Google Scholar]
- Taneda, T. , Miyata, S. , Kousaka, A. , Inoue, K. , Koyama, Y. , Mori, Y. , & Tohyama, M. (2007). Specific regional distribution of protein arginine methyltransferase 8 (PRMT8) in the mouse brain. Brain Research, 1155, 1–9. [DOI] [PubMed] [Google Scholar]
- Temple, S. , & Qian, X. (1996). Vertebrate neural progenitor cells: subtypes and regulation. Current Opinion in Neurobiology, 6(1), 11–17. [DOI] [PubMed] [Google Scholar]
- Terai, K. , Matsuo, A. , & McGeer, P. L. (1996). Enhancement of immunoreactivity for NF‐kappa B in the hippocampal formation and cerebral cortex of Alzheimer's disease. Brain Research, 735(1), 159–168. [DOI] [PubMed] [Google Scholar]
- Teyra, J. , Huang, H. , Jain, S. , Guan, X. , Dong, A. , Liu, Y. , Tempel, W. , Min, J. , Tong, Y. , Kim, P. M. , Bader, G. D. , & Sidhu, S. S. (2017). Comprehensive analysis of the human SH3 domain family reveals a wide variety of non‐canonical specificities. Structure, 25(10), 1598–610.e3. [DOI] [PubMed] [Google Scholar]
- Toth, R. P. , & Atkin, J. D. (2018). Dysfunction of optineurin in amyotrophic lateral sclerosis and glaucoma. Frontiers in Immunology, 9, 1017‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière, Hélène , Chebli, K. , Zekri, L. , Courselaud, B. , Blanchard, J. M. , Bertrand, E. , & Tazi, J. (2003). The RasGAP‐associated endoribonuclease G3BP assembles stress granules. Jcell Biol, 160(6), 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai, W.‐C. , Gayatri, S. , Reineke, L. C. , Sbardella, G. , Bedford, M. T. , & Lloyd, R. E. (2016). Arginine demethylation of G3BP1 promotes stress granule assembly. The Journal of Biological Chemistry, 291(43), 22671–22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, W. C. , Reineke, L. C. , Jain, A. , Jung, S. Y. , & Lloyd, R. E. (2017). Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule‐nucleating protein G3BP1. The Journal of Biological Chemistry, 292(46), 18886–18896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushev, G. , Glock, C. , Heumuller, M. , Biever, A. , Jovanovic, M. , & Schuman, E. M. (2018). Alternative 3' UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron, 98(3), 495–511.e6. [DOI] [PubMed] [Google Scholar]
- Umeda, Y. , Taniguchi, S. , Arima, K. , Piao, Y.‐S. , Takahashi, H. , Iwatsubo, T. , Mann, D. , & Hasegawa, M. (2004). Alterations in human tau transcripts correlate with those of neurofilament in sporadic tauopathies. Neuroscience Letters, 359(3), 151–154. [DOI] [PubMed] [Google Scholar]
- Valerio, A. , Boroni, F. , Benarese, M. , Sarnico, I. , Ghisi, V. , Bresciani, L. G. , Ferrario, M. , Borsani, G. , Spano, P. , & Pizzi, M. (2006). NF‐kappaB pathway: a target for preventing beta‐amyloid (Abeta)‐induced neuronal damage and Abeta42 production. European Journal of Neuroscience, 23(7), 1711–1720. [DOI] [PubMed] [Google Scholar]
- Ververis, A. , Dajani, R. , Koutsou, P. , Aloqaily, A. , Nelson‐Williams, C. , Loring, E. , Arafat, A. , Mubaidin, A. F. , Horany, K. , Bader, M. B. , & Al‐Baho, Y. (2020). Distal hereditary motor neuronopathy of the Jerash type is caused by a SIGMAR1 c. 500A> T missense mutation. Journal of Medical Genetics, 57(3), 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vognsen, T. , Møller, I. R. , & Kristensen, O. (2013). Crystal structures of the human G3BP1 NTF2‐like domain visualize FxFG Nup repeat specificity. PLoS One, 8(12), e80947‐e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.‐J. , Liu, C. , Shan, K. , Liu, B.‐H. , Li, X.‐M. , Zhang, S.‐J. , Zhou, R.‐M. , Dong, R. , Yan, B. , & Sun, X.‐H. (2018). Circular RNA‐ZNF609 regulates retinal neurodegeneration by acting as miR‐615 sponge. Theranostics, 8(12), 3408–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Fu, D. , Chen, Y. , Su, J. , Wang, Y. , Li, X. , Zhai, W. , Niu, Y. , Yue, D. , & Geng, H. (2018). G3BP1 promotes tumor progression and metastasis through IL‐6/G3BP1/STAT3 signaling axis in renal cell carcinomas. Cell Death & Disease, 9(5), 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshhans, K. , & Bassell, G. J. (2011). Netrin‐1‐induced local beta‐actin synthesis and growth cone guidance requires zipcode binding protein 1. Journal of Neuroscience, 31(27), 9800–9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. P. , Cardenas, A. M. , Marissen, W. E. , & Lloyd, R. E. (2007). Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell HostMicrobe, 2(5), 295–305. [DOI] [PubMed] [Google Scholar]
- White, J. P. , & Lloyd, R. E. (2011). Poliovirus unlinks TIA1 aggregation and mRNA stress granule formation. Jvirol, 85(23), 12442–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, D. , Li, K. W. , Zheng, J. Q. , Chang, J. H. , Smit, A. , Kelly, T. , Merianda, T. T. , Sylvester, J. , Van Minnen, J. , & Twiss, J. L. (2005). Differential transport and local translation of cytoskeletal, injury‐response, and neurodegeneration protein mRNAs in axons. Journal of Neuroscience, 25(4), 778–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin, B. , & Ivanov, P. (2019). Stress granules and neurodegeneration. Nature Reviews Neuroscience, 20(11), 649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Wang, P. , Liu, Y. , Zhang, Y. , Fan, W. , Upton, M. P. , Lohavanichbutr, P. , Houck, J. R. , Doody, D. R. , Futran, N. D. , Zhao, L. P. , Schwartz, S. M. , Chen, C. , & Méndez, E. (2013). Integrative genomics in combination with RNA interference identifies prognostic and functionally relevant gene targets for oral squamous cell carcinoma. PLoS Genetics, 9(1), e1003169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P. , Mathieu, C. , Kolaitis, R.‐M. , Zhang, P. , Messing, J. , Yurtsever, U. , Yang, Z. , Wu, J. , Li, Y. , Pan, Q. , Yu, J. , Martin, E. W. , Mittag, T. , Kim, H. J. , & Taylor, J. P. (2020). G3BP1 Is a tunable switch that triggers phase separation to assemble stress granules. Cell, 181(2), 325–45.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C.‐Y. , & Kuo, H.‐C. (2019). The emerging roles and functions of circular RNAs and their generation. Journal of Biomedical Science, 26(1), 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste, R. (2011). Dendritic spines and distributed circuits. Neuron, 71(5), 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri, L. , Chebli, K. , Tourrière, Hélène , Nielsen, F. C. , Hansen, T. V. O. , Rami, A. , & Tazi, J. (2005). Control of fetal growth and neonatal survival by the RasGAP‐associated endoribonuclease G3BP. Molecular and Cellular Biology, 25(19), 8703–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Ma, Y. , Zhang, S. , Liu, H. , He, H. , Li, N. , Gong, Y. , Zhao, S. , Jiang, J.‐D. , & Shao, R.‐G. (2015). Involvement of Ras GTPase‐activating protein SH3 domain‐binding protein 1 in the epithelial‐to‐mesenchymal transition‐induced metastasis of breast cancer cells via the Smad signaling pathway. Oncotarget, 6(19), 17039–17053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L.‐N. , Zhao, L. , Yan, X.‐L. , & Huang, Y.‐H. (2019). Loss of G3BP1 suppresses proliferation, migration, and invasion of esophageal cancer cells via Wnt/β‐catenin and PI3K/AKT signaling pathways. Journal of Cellular Physiology, 234(11), 20469–20484. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Sharma, N. R. , Zheng, Z.‐M. , & Chen, M. (2019). Viral regulation of RNA granules in infected cells. Virologica Sinica, 34(2), 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. E. , Sloan, S. A. , Clarke, L. E. , Caneda, C. , Plaza, C. A. , Blumenthal, P. D. , Vogel, H. , Steinberg, G. K. , Edwards, M. S. B. , Li, G. , Duncan, J. A. , Cheshier, S. H. , Shuer, L. M. , Chang, E. F. , Grant, G. A. , Gephart, M. G. H. , & Barres, B. A. (2016). Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron, 89(1), 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Hartmann, H. , Minh Do, V. , Abramowski, D. , Sturchler‐Pierrat, C. , Staufenbiel, M. , Sommer, B. , van de Wetering, M. , Clevers, H. , Saftig, P. , De Strooper, B. , He, X. I. , & Yankner, B. A. (1998). Destabilization of β‐catenin by mutations in presenilin‐1 potentiates neuronal apoptosis. Nature, 395(6703), 698–702. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Liu, S. , Liu, G. , Oztürk, A. , & Hicks, G. G. (2013). ALS‐associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genetics, 9(10), e1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]


