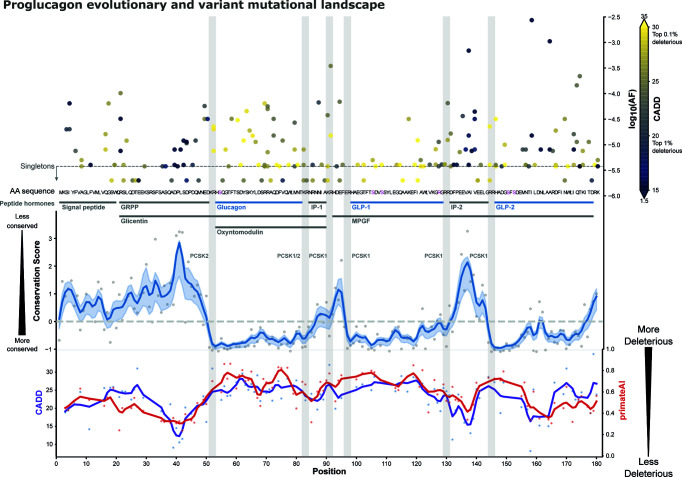
Figure 2.
Proglucagon mutational landscape. Functionally described peptide hormones are highlighted (blue), which are more conserved in an evolutionary trace analysis employing Rate4Site scores [blue line, gaussian CI: 50% (46, 47)], than other parts of the precursor and peptides such as IP1/IP2, GRPP, and the signal peptide (most conserved position has a score of –1). All 184 missense variants are displayed along their predicted CADD scores (color-grading) (48) and their corresponding max allele frequencies (top-right y-axis). Peptide cleavage motifs are highlighted as grey bars alongside their known enzymes. Post-translational modification sites are highlighted (pink) in the amino acid sequence. Predicted CADD (purple) and primateAI (red) scores are presented (bottom curves) (48, 49), indicating higher mean predicted deleteriousness in glucagon and GLP-1. See SF1 for an interactive version.