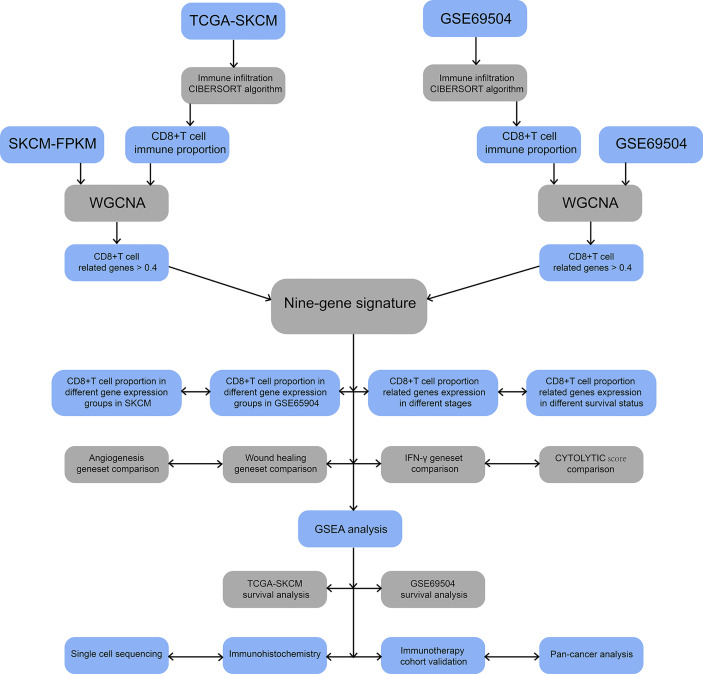
Figure 1.
Flowchart for identification of co-expressed genes promoting CD8+ T cells infiltration. SKCM-FPKM contains 470 melanoma cancer tissue samples. GSE65904 contains 214 melanoma samples. CIBERSORT algorithm was applied to calculate the infiltration of CD8+ T cells in melanoma tissue samples from two datasets. WGCNA was used to generate a co-expressed gene network, and the two datasets were intermingled to obtain nine essential genes. The main pathways of critical gene enrichment were obtained in GSEA analysis. Survival analysis was used to determine the influence of critical genes on the outcome. Single-cell sequencing and immunohistochemical experiments confirmed that the expression of these genes was high in CD8+ T cells. The comparison between 9-gene signature with angiogenesis geneset, wound healing geneset, immune response geneset and IFN-γ genset were analyzed. Follow-up cohorts with immunotherapy were also used to validate the role of critical genes in immunotherapy. The 9-gene signature that related to CD8+ T cell infiltration in other cancers were verified.