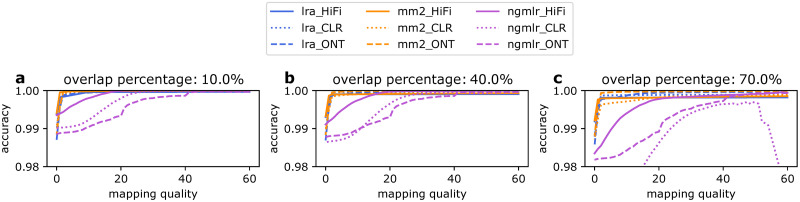
Fig 1. Mapping accuracy.
Mapping accuracy of lra, minimap2 and ngmlr on simulated HiFi, CLR reads for lengths between 5–50kb and ONT reads for lengths between 1–50kb. Simulated reads were mapped to genome GRCh38. A read is considered as correctly mapping if the reported mapping interval has ≥ 10%, 40%, 70% overlap with the truth interval. paftools.js mapeval was used to evaluate the mapping accuracy.