Figure 1.
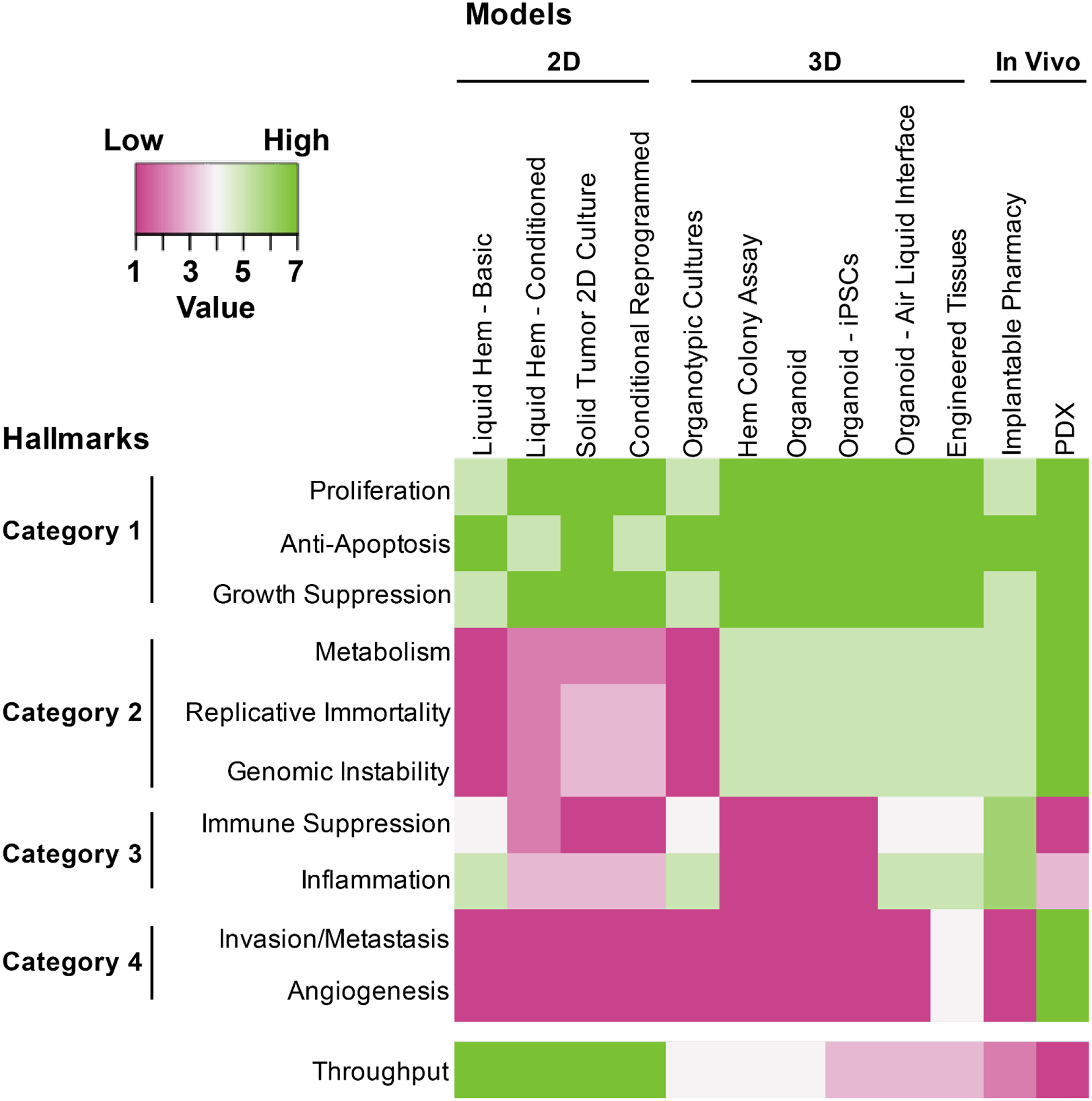
The 10 hallmarks of cancer are organized into 4 categories based on feasibility of various ex vivo platforms to accurately assess sensitivity to drugs targeting those hallmarks. In Category 1, most platforms are highly capable of directly assessing cell proliferation and apoptosis or, at least, assessing biomarkers of these phenotypic states. The Category 2 hallmarks of metabolism, replicative immortality, and genomic instability are better modeled with approaches that enable robust cell proliferation and/or that can readout phenotypes beyond short term cultures. Hallmarks in Category 3 can be best modeled with assays that include heterogeneous cell mixtures that are derived from or mimic the native tumor state. In this way, some of the basic models may actually perform better than models with greater perturbations on the tumor biopsy, however, these heterogeneous cell mixtures may be added back with some of the sophisticated engineered models. Finally, Category 4 hallmarks may be the most challenging, but can be modeled using bioengineered 3-dimensional approaches or patient-derived xenograft models. In general, the 3-dimensional and in vivo approaches are able to accurately model a larger number of hallmarks, though it is worth noting that these strategies currently do not have the throughput of the more basic approaches.
