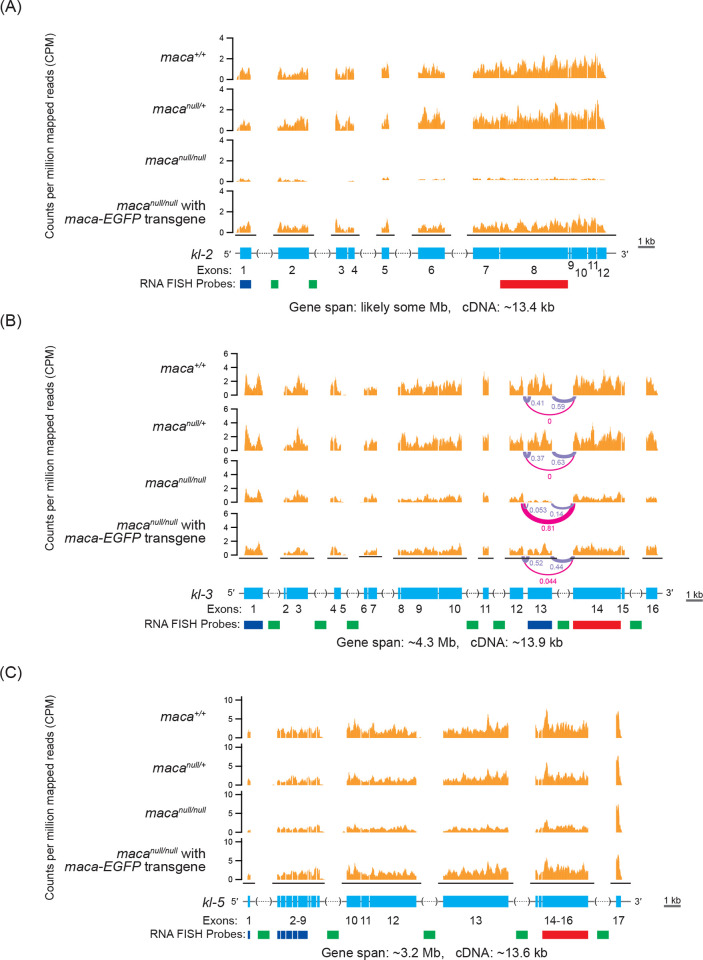
Fig 6. Poly-A+ RNA-seq read mapping counts for kl-2, kl-3, and kl-5 mRNAs show their reduction in macanull/null testes.
Poly-A+ RNA-seq normalized read counts (counts per million mapped reads, CPM) in the (A) kl-2, (B) kl-3, and (C) kl-5 gene regions. For kl-3, splicing ratios among (1) exon 12—exon 13, (2) exon 13—exon 14, and (3) exon 12—exon 14 (= exon 13-skipped) determined by LeafCutter [39] are shown. Only one replicate out of 3 is shown for each genotype. All replicates are shown in S5–S7 Figs. Gene structures are shown with exons (cyan), introns (black line), intronic satellite DNA repeats (dashed line in parentheses. Contain gaps in the publically available Drosophila genome sequence). Regions targeted by RNA FISH probes used in Figs 8, 10, S10, S13, S14, S15 and S16 (blue, green, and red).