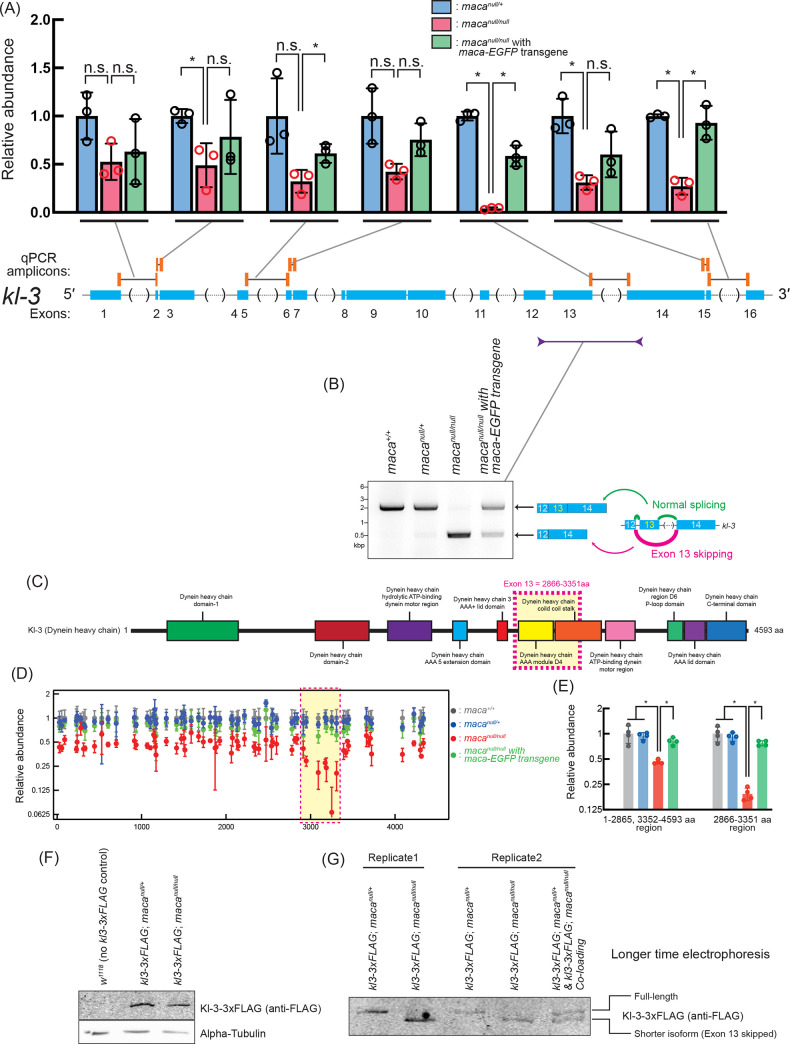
Fig 9. kl-3 mRNA reduction and exon 13 skipping in macanull/null testes revealed by RT-(q)PCR.
(A) Quantification of kl-3 transcripts in testis RNA by RT-qPCR. The RT step was performed with random hexamer primers. qPCR amplicons targeted by designed primer sets are indicated by orange bars. The primers span exon-exon junctions to amplify only spliced transcripts. Black lines indicate intron regions not included in the amplicons. Data were normalized to actin5C mRNA and the mean of macanull/+. Mean +/- SD (n = 3 biological replicates). P-values <0.05 (Student’s t-test, unpaired, two-tailed) are indicated by *. (B) Agarose gel electrophoresis of RT-PCR products using the indicated primers to examine kl-3 exon 13 skipping in testis RNA. (C) Diagram of Kl-3 protein domains. Amino acid residues 2866–3351 corresponding to the exon 13 is indicated with a magenta dash-lined yellow box. (D) Abundance of peptide fragments derived from Kl-3 protein determined by mass spec with TMT labeling plotted along the Kl-3 amino acid residue numbers. Data were normalized to the means of maca+/+. Mean +/- SD (n = 4 biological replicates). The amino acid residues 2866–3351 corresponding to the kl-3 mRNA exon 13 is indicated with a magenta dash-lined yellow box. (E) Abundance of Kl-3 protein determined using peptide fragments data (left) from the 1–2865 or 3352–4593 aa region or (right) from the 2866–3351 aa region, of Kl-3 in mass spec with TMT labeling. Data were normalized to the means of maca+/+. Mean +/- SD (n = 4 biological replicates). P-values <0.05 (Student’s t-test, unpaired, two-tailed) are indicated by *. (F, G) Western blots of testis lysates for Kl-3-3xFLAG using (F) a standard protocol and (G) an extended SDS-PAGE gel electrophoresis time to achieve better separation. The FLAG tag was inserted at the C-terminal end of Kl-3 at the endogenous kl-3 locus on the Y-chromosome [40].