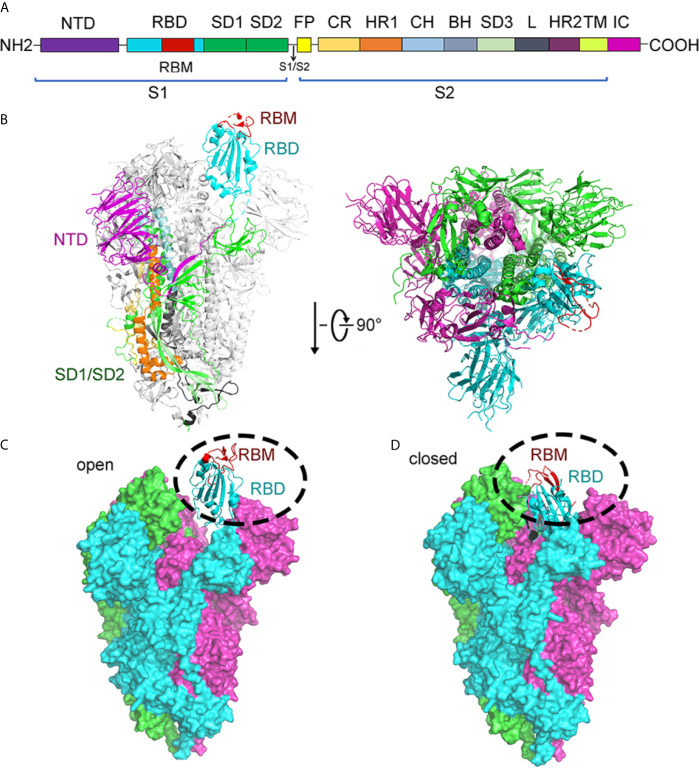
Figure 6.
Structure of SARS-CoV-2 spike protein. (A) Schematic of the SARS-CoV-2 spike protein primary structure colored by domain. NTD, N-terminal domain; RBD, receptor binding domain; RBM, receptor binding motif; SD1, subdomain 1; SD2, subdomain 2; FP, fusion peptide; CR, connecting region; HR1, heptad repeat 1; CH, central helix; BH, β-hairpin; SD3, subdomain 3; HR2, heptad repeat 2; TM, transmembrane domain; IC, cytoplasmic tail. Arrows denote protease cleavage sites. (B) From the side view and top view, the Cryo-EM structure of the SARS-CoV-2 spike in the prefusion conformation shows that a single RBD is in the up conformation and other RBDs are in the down conformation in either white or gray. The RBD, which in up conformation, is shown in ribbons colored corresponding to the schematic in (A). (C) Cryo-EM structure of the SARS-CoV-2 spike in the open state. There is one open RBD domain. The three chains are depicted in green, cyan, and magnet, respectively. RBD and RBM are shown in cyan and red cartoon, respectively. (D) Cryo-EM structure of the SARS-CoV-2 Spike in the closed state. The three chains are depicted in green, cyan, and magnet, respectively. RBM is colored in red. RBD and RBM are shown in cyan and red cartoon, respectively. All structures are drawn by Pymol.