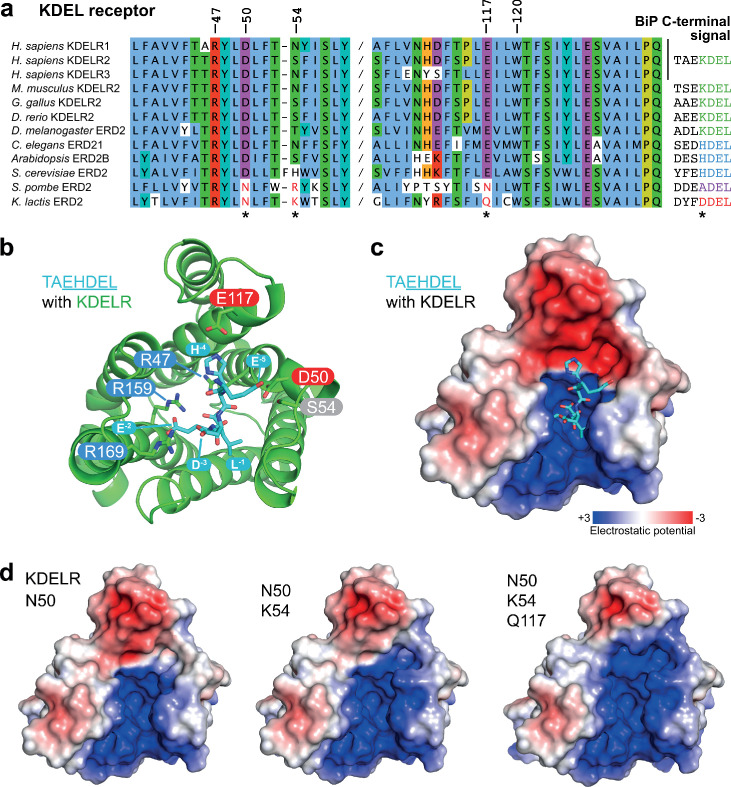
Figure 5. Charge distribution across the luminal entrance to the KDEL receptor binding pocket.
(a) KDEL receptor sequence alignment showing two regions centred around amino acid D50 and W120 of the human proteins. Cognate retrieval signal variants are shown to the right of the alignment. (b) The structure of the KDEL receptor with bound TAEHDEL highlighting key residues involved in ligand binding and variant residues D50, N54, and E117. (c) The charged surface for the WT KDEL receptor and (d) N50, N50/K54 and N50/K54/Q117 mutants is shown.