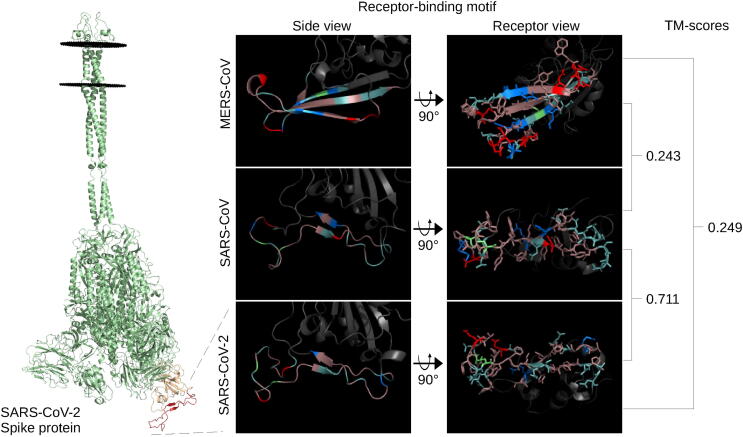
Fig. 1.
Spike receptor-binding motif comparison. The full-length SARS-CoV-2 spike protein (green), left, modelled using ProtCHOIR is shown with the receptor-binding domain (yellow) and receptor-binding motif (red) marked. The RBMs from the side view are shown, middle, with the amino acids labelled by color: red for acidic (D,E), blue for basic (H,R,K), light teal for polar non-charged (S,N,T,Q), dirty violet for hydrophobic (A,V,I,L,M,F,W,P,G,Y), and lime green for cysteine residues. RBMs from the host cell receptor side are shown, right, with amino acid stick configurations. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)