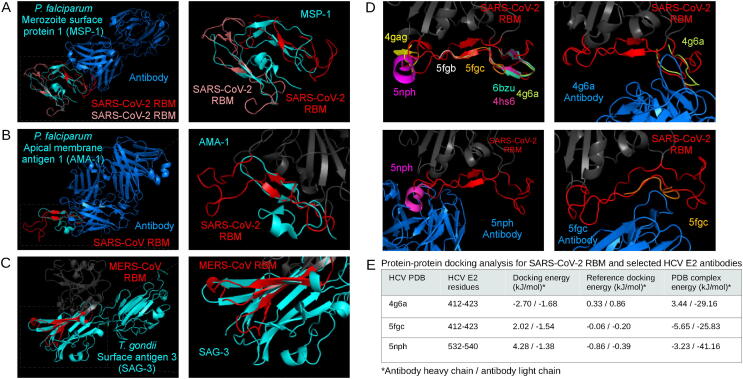
Fig. 4.
Analysis of exogenous structural alignments. Plasmodium falciparum merozoite surface protein 1 (A) and apical membrane antigen 1 (B) structurally aligned with the SARS-CoV-2 RBM and SARS-CoV RBM, respectively. The Toxoplasma gondii surface antigen 3 aligned with the MERS-CoV RBM (C). RBMs are labelled red, mimicked proteins are cyan, and potential interaction partners are marine blue (A, B, C). Hepatitis C virus epitopes structurally aligned to the SARS-CoV-2 RBM, and the respective antibody structures from PDBs 5fgc, 5nph, 4g6a docked to the RBM (D) using ClusPro PIPER “antibody” mode. Interaction energy scores predicted using FoldX on docked and experimental complexes (E). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)