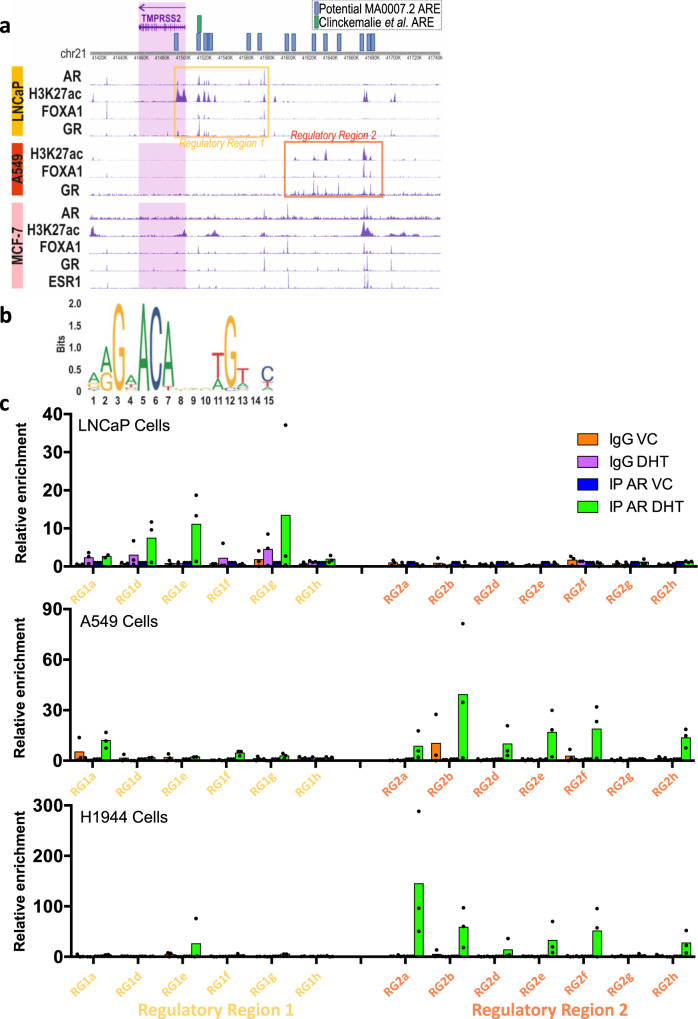
Fig. 6. Potential regulatory regions in the TMPRSS2 promoter.
a ChIP-sequencing peaks of AR, FOXA1, GR and H3K27ac in LNCaP cells; FOXA1, GR and H3K27ac in A549 lung cells; AR, H3K27ac, FOXA1, GR, ESR1 in MCF-7 breast cancer cells. The TMPRSS2 gene is highlighted in the purple shaded box and the potential regulatory region 1 is boxed in yellow, the potential regulatory region 2 is boxed in orange. Potential AREs are marked in blue boxes (MA0007.2) or green (determined by24). Position of potential MA0007.2 motifs around the TMPRSS2 gene separated into two regions, the first region (regulatory region 1) covering areas of TSS and promoter/enhancers, the second region (regulatory region 2) covers more distant enhancer regions. b AR motif MA0007.2 from JASPAR curated motif database. c ChIP-qPCR of AR binding, and IgG control, to AR-motif-containing sites within regulatory regions 1 and 2 in LNCaP, A549, and H1944 cells treated with 10 nM dihydrotestosterone (DHT) or vehicle control (VC) for 4 h. Data are normalised to input and made relative to AR-IP VC. Data are the mean of three independent repeats. A maximum of one outlier was removed from each treatment using Grubbs’ test (alpha = 0.05). Source data are provided as a Source Data file.