FIGURE 1.
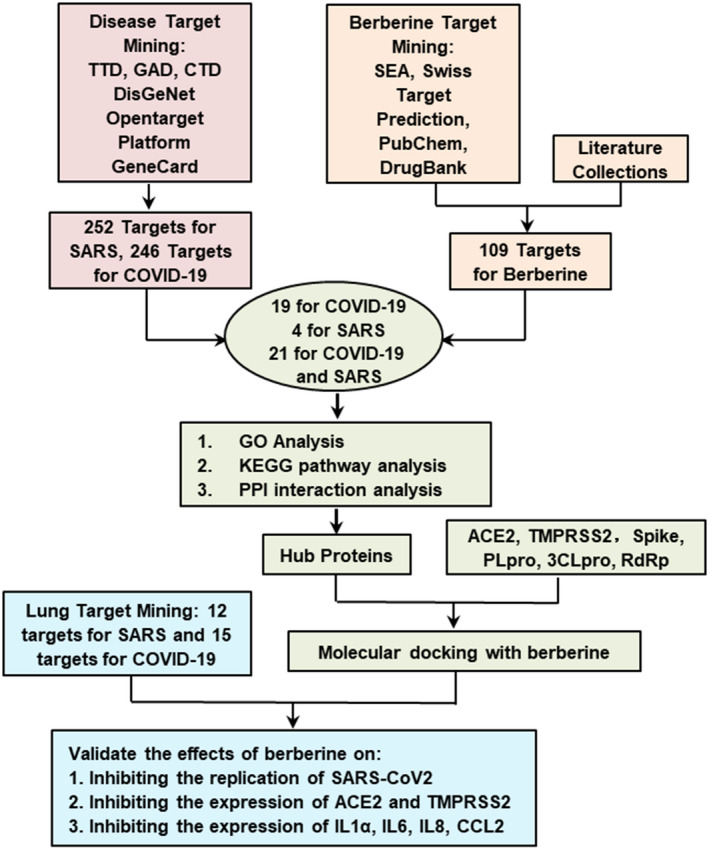
The workflow of computational modeling used for analysis of berberine as a promising candidate against COVID‐19 and SARS. TTD: therapeutic target database; GAD: genetic association database; SEA: similarity ensemble approach; PPI: protein‐protein interactions; ACE2: angiotensin‐converting enzyme 2; TMPRSS2: transmembrane serine protease 2; PLpro: Papain‐like Protease; 3CLpro: coronavirus main proteinase; RdRp: RNA‐dependent RNA polymerase. First, a total of 254 genes for SARS and 247 genes for COVID‐19 were selected as possible targets for berberine. Separately, 109 inflammatory and biological targets of berberine were collected based on literatures and following published databases: hitpick, swiss target prediction, SEA, pubchem, and drugbank. Mining berberine targets onto identified disease targets uncovers that berberine might potentially regulate 21 targets for both COVID‐19 and SARS, 4 targets for SARS specifically, and 19 targets for COVID‐19 specifically. With biological targets of berberine for the prevention and treatment of COVID‐19 and SARS established, GO, KEGG pathway, and PPI analysis were conducted to uncover the mechanistic details of berberine regulation in key pathways. Moreover, hub host proteins are determined for further molecular docking analysis. Combining crucial virus proteins and vital host receptor proteins in virus infection and duplication process, molecular docking was further applied to investigate the possible binding modes of berberine to these targets. Based on the lung target mining and molecular docking results, the inhibition of berberine on ACE2, TMPRSS2, IL1α, IL6, IL8, and CCL2 were further validated
