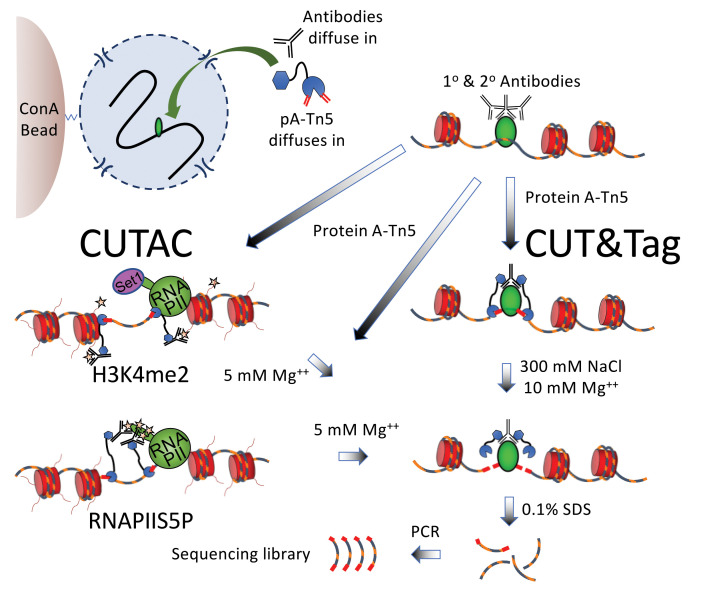
Figure 2. Scheme for simultaneous CUT&Tag and (H3K4me2 or RNAPIIS5P) CUTAC.
CUT&Tag-direct is performed in situ in single PCR tubes with Concanavalin A (ConA) bead-bound nuclei that remain intact throughout the protocol during successive liquid changes, incubations and washes, 12 cycles of PCR amplification, and one SPRI bead cleanup. CUTAC is performed identically except that low-salt conditions are used for tagmentation. H3K4me2 CUTAC maps accessible sites near H3K4me2/3-marked (starred) nucleosome tails, which are methylated by the conserved Set1 lysine methyltransferase. The complex that includes Set1 associates with the initiation form of RNAPII, which is heavily phosphorylated on Serine-5 of the heptameric C-terminal domain repeat units on the largest RNAPII subunit (RNAPIIS5P). For RNAPIIS5P CUTAC, pA-Tn5 is anchored directly to RNAPIIS5 phosphates (starred). Whereas CUT&Tag is suitable for any chromatin epitope, CUTAC is specific for H3K4me2, H3K4me3, and RNAPIIS5P. The only other difference between the protocols is that tagmentation is performed in the presence of 300 mM NaCl for CUT&Tag and in a low ionic strength buffer for CUTAC.