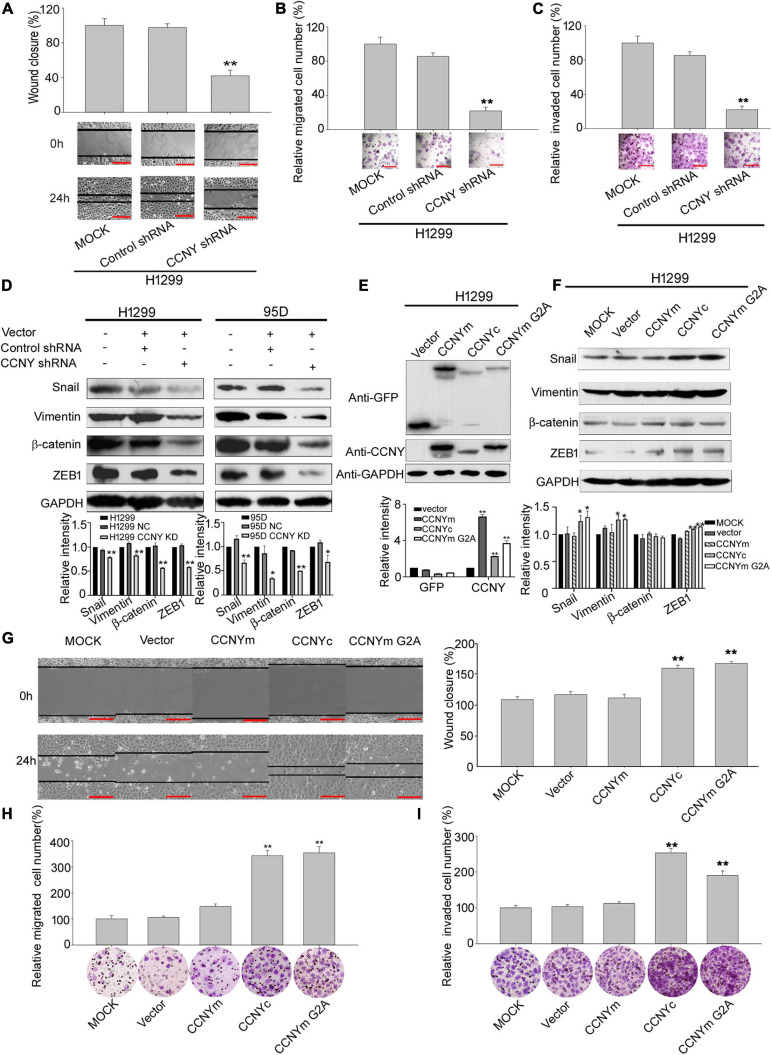
FIGURE 3.
CCNYc could promote cell motility and invasiveness in H1299 cells. (A) Wound healing assay performed with H1299-CCNY KD and control cells over 24 h. The migration activity is shown as mean ± SEM. Bars represent 200 μm. **p < 0.01 vs. NC cells (t-test, N = 3). (B,C) Transwell assay performed with H1299-CCNY KD and the control cells. Cells migrated or invaded through the membrane were calculated and standardized. Bars represent 200 μm. **p < 0.01 vs. NC cells (t-test, N = 3). (D) The upper panel, levels of β-catenin, vimentin, ZEB1 and snail were measured by WB; the lower panel, the level of proteins was quantified by gray analysis. *p < 0.05 vs. NC cells, **p < 0.01 vs. NC cells (t-test). (E) Expression levels of GFP and CCNY were examined by immunoblotting. The levels of GFP and CCNY were quantified by gray analysis in the lower panel. **p < 0.01 vs. H1299- pEGFP-N1 cells (t-test). (F) Expression of ZEB1, vimentin, β-catenin and snail were examined by WB. The quantification of the proteins was listed in the lower panel. *p < 0.05 vs. H1299-vector cells, **p < 0.01 vs. H1299-pEGFP-N1 cells (t-test). (G) Wound healing assay performed with CCNYm, CCNYc, and CCNYm G2A upregulation cells. The migration activity is expressed as mean ± SEM. **p < 0.01 vs. H1299 cells transfected with pEGFP-N1 vector (t-test, N = 3). (H,I) Transwell migration assay and invasion assay were used. Bars represent 200 μm. The data are shown as mean ± SEM. **p < 0.01 vs. H1299 cells transfected with pEGFP-N1 vector (t-test, N = 3).