Figure 1.
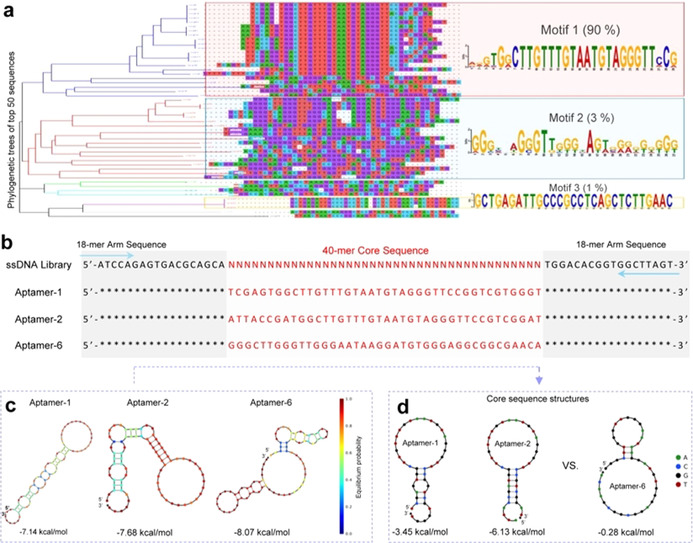
Aptamer sequences specific for viral S/RBD. a) Aptamers were developed using a target‐based enrichment process and final products were sequenced. The top 50 predominant aptamer sequences were selected from over 100 000 reads. Phylogenetic tree analysis identified three well‐preserved sequence motifs. b) Aptamers‐1 and ‐2 were derived from the Motif 1 sequence and aptamer‐6 from the Motif 2 sequence. Sequences of the ssDNA library used for aptamer development are also shown. Blue arrows indicate 18‐mer consistent arm sequences at both ends for primer annealing of PCR amplification. Red lettering indicates central cores composed of 40‐mer specific sequences of the aptamers or random sequences in the Random ssDNA library. c) Minimum free energy secondary structures of aptamers‐1, ‐2, and ‐6. d) Core sequence comparison confirms different secondary structures for aptamers derived from Motif 1 and 2 sequences.
