FIGURE 1.
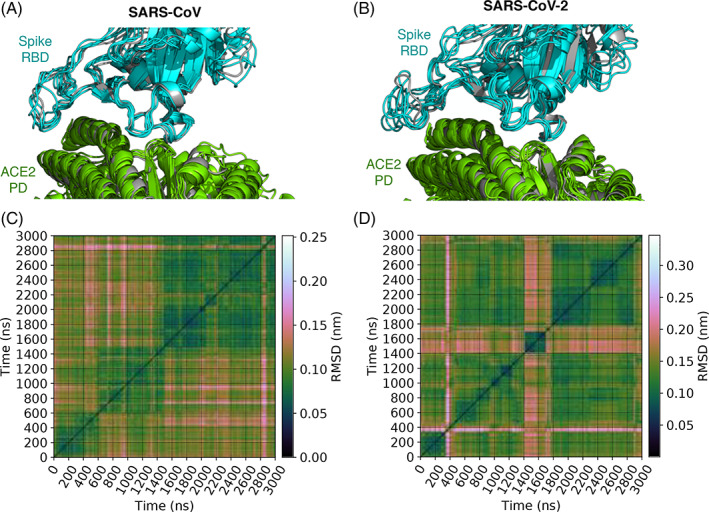
Binding modes of spike‐angiotensin converting enzyme 2 (ACE2) complexes in MD simulations. A and B show, respectively, the distinct binding mode conformations of the complexes containing spike proteins of CoV and CoV‐2. These conformations are superimposed over the X‐ray structures of spike‐ACE2 complexes (gray)—ACE2 bound with CoV spike is compared against 2AJF, 13 and ACE2 bound to CoV‐2 spike is compared against 6M0J. 7 They are the lowest energy conformations of the five and six binding modes identified, respectively, for the complexes involving CoV and CoV‐2 spike proteins. Binding modes are identified by clustering conformations extracted every nanosecond from MD. Conformational clustering is performed using affinity propagation 30 , 32 in which we take RMSD as an index of similarity between conformations. C and D show these pairwise RMSDs
