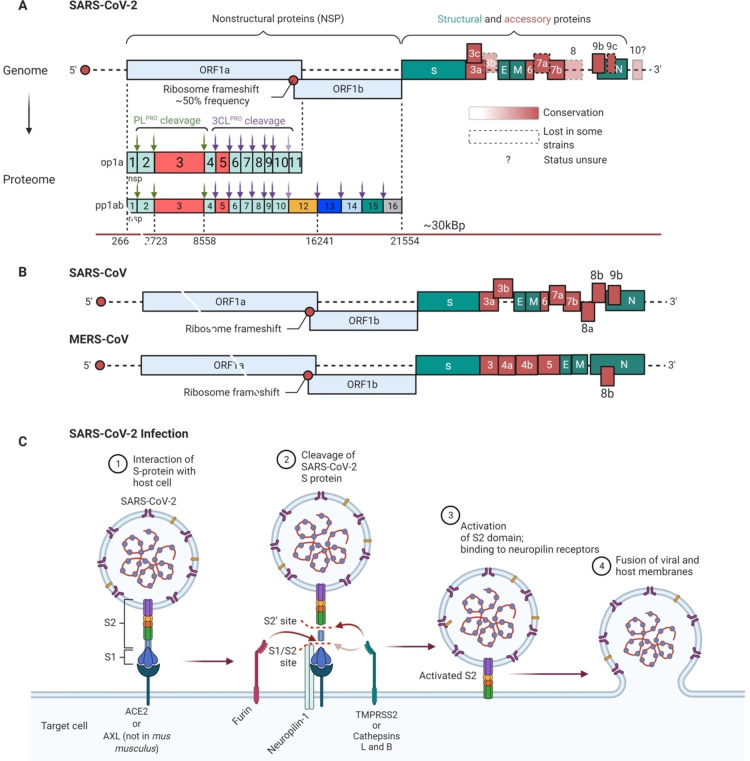
Figure 2.
A brief guided tour of CoV biology. (A) SARS‐CoV2 genome and proteolytic processing. In the genome, light blue denotes ORF1ab; green denotes structural proteins; red ancillary proteins. Shading denotes conservation; dotted borders indicates variants lacking this gene are known (truncations in ORF7b are known, but not shown here)[331]. In the protein, Pp1a and pp1ab denotes different gene products derived from ORF1ab. Height of boxes denotes translation rate. Note NSP11 is only 13 amino acids in length; other NSPs range from 180 amino acids (NSP1) to 1945 amino acids (NSP3). [371] For NSPs, red boxes indicate proteases; Green down arrow indicates a step catalyzed by PLPRO; purple downwards arrow indicates cleavage by 3CLPRO (faded arrows indicate not necessary for replication in some CoVs, but can lead to attenuation). [372] Red line indicates genomic positions. (B) Genomes of SARS‐CoV and MERS‐CoV for comparison. (C) SARS‐CoV‐2 infection occurs when the S‐protein binds to an acceptor protein on the cell surface. This protein is believed to be ACE‐2 or AXL (except in mice). Upon formation of the complex, the S‐protein is cleaved by several different proteases (Furin and TMPRSS2 or cathepsins L or B). Other proteins, such as neuropilin‐1 are required for viral entry. Upon cleavage the activated S‐protein can orchestrate fusion with the plasma membrane and delivery of the genome to the cell.