Figure 3.
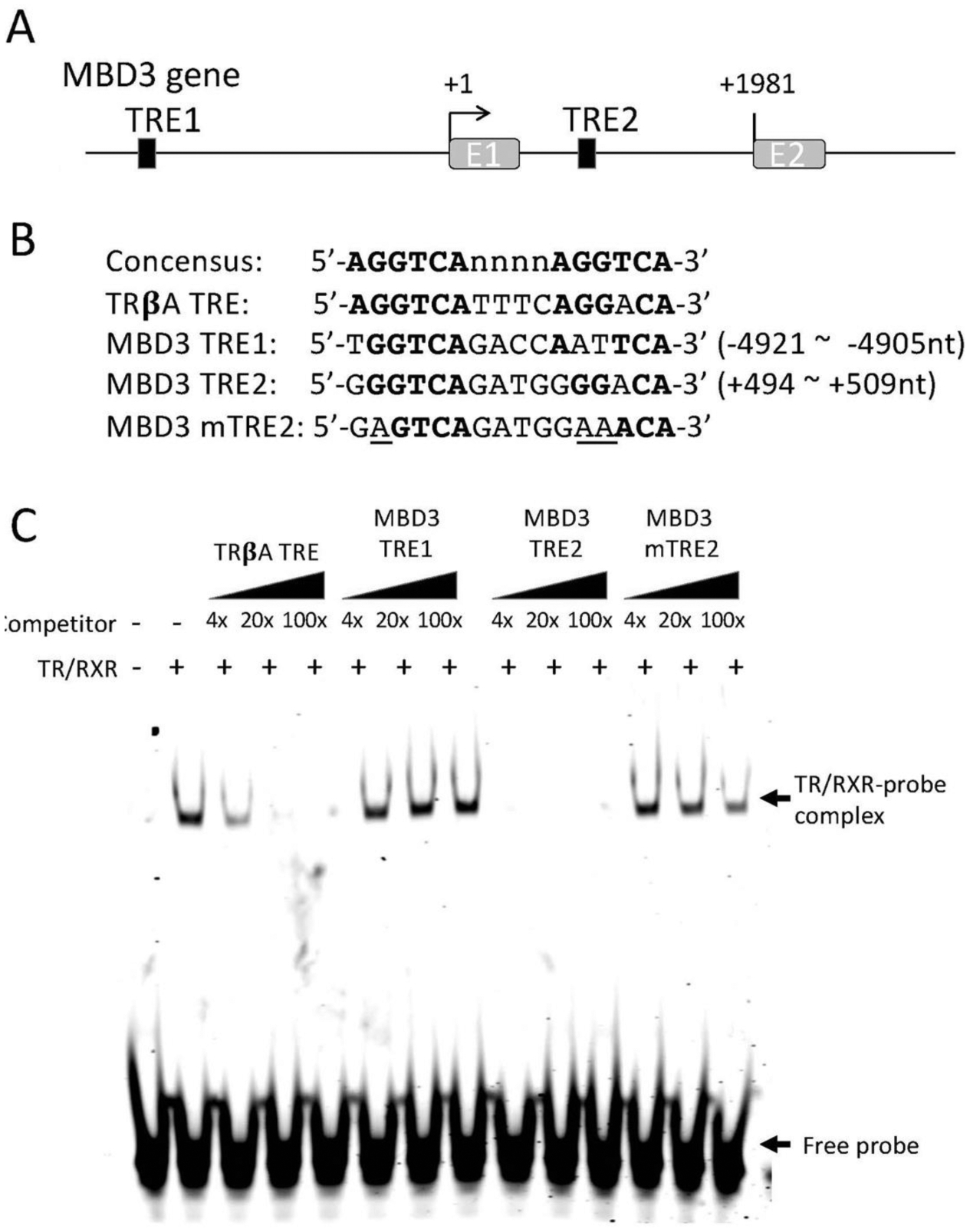
TR binds to a putative TRE within the first intron of the MBD3 gene in vitro. (A) Schematic diagram showing two putative TREs (solid boxes) in X. tropicalis MBD3 gene. (B) The sequences of the putative TREs are shown in comparison with the sequences of consensus TRE and the TRE of Xenopus laevis TRβA gene (TRβA TRE). The numbers indicate the locations relative to the transcriptional start site of MBD3 gene based on the 5’-terminus of the reported MBD3 cDNA sequence (denoted as +1). A mutant version of MBD3 TRE2 (MBD3 mTRE2, with the mutated nucleotides underlined) was made for TR/RXR binding assay in comparison with the wild type TREs. Bold letters indicate nucleotides conserved with the consensus TRE. Note that the putative TRE1 (MBD3 TRE1) is in the 5’-proximity of its promoter and the putative TRE2 (MBD3 TRE2) is in the first intron of the gene. E1: exon1; E2: exon2. (C) The putative MBD3 TRE2 but not TRE1 compete for binding to TR/RXR heterodimers against labeled TRβA TRE in a gel mobility shift assay. The labelled double stranded probe containing TRβA TRE was mixed with in vitro translated TR/RXR heterodimers in the presence or absences of 4x, 20x, or 100x of unlabelled wild type or mutant MBD3 TREs as indicated. The reaction mixtures were analyzed by electrophoretic mobility shift assay (EMSA). The mock in vitro translation mixture was used as a negative control for TR/RXR heterodimers (lane 1 on the left). The locations of TR/RXR bound probe (TR/RXR-probe complex) and unbound probe (Free probe) are indicated with arrows. Note that MBD3 TRE1 had little competition against the labelled TRβA TRE probe while MBD3 TRE2 strongly competed against the labelled probe. Mutations in the MBD3 TRE2 drastically reduced its ability to compete against the labeled probe for TR/RXR binding.
