Summary
Pre‐harvest sprouting (PHS), the germination of grain before harvest, is a serious problem resulting in wheat yield and quality losses.
Here, we mapped the PHS resistance gene PHS‐3D from synthetic hexaploid wheat to a 2.4 Mb presence–absence variation (PAV) region and found that its resistance effect was attributed to the pleiotropic Myb10‐D by integrated omics and functional analyses.
Three haplotypes were detected in this PAV region among 262 worldwide wheat lines and 16 Aegilops tauschii, and the germination percentages of wheat lines containing Myb10‐D was approximately 40% lower than that of the other lines. Transcriptome and metabolome profiling indicated that Myb10‐D affected the transcription of genes in both the flavonoid and abscisic acid (ABA) biosynthesis pathways, which resulted in increases in flavonoids and ABA in transgenic wheat lines. Myb10‐D activates 9‐cis‐epoxycarotenoid dioxygenase (NCED) by biding the secondary wall MYB‐responsive element (SMRE) to promote ABA biosynthesis in early wheat seed development stages.
We revealed that the newly discovered function of Myb10‐D confers PHS resistance by enhancing ABA biosynthesis to delay germination in wheat. The PAV harboring Myb10‐D associated with grain color and PHS will be useful for understanding and selecting white grained PHS resistant wheat cultivars.
Keywords: Aegilopstauschii, functional analyses, grain color, integrated omics, presence–absence variation, pre‐harvest sprouting, synthetic wheat, Triticumaestivum
Introduction
Common wheat (Triticum aestivum, AABBDD) is a staple food crop for more than one‐third of the global human population and provides c. 20% of the calories consumed by humans globally (Shewry & Hey, 2015). The availability of reference sequences and the rise of the pangenome project of wheat have allowed the identification of genes and their structural variations that involve the single‐nucleotide polymorphisms (SNPs) and the loss of genomic regions encoding protein‐coding genes in some individuals (i.e. presence–absence variation, PAV) (Uauy, 2017; Jia et al., 2018; Walkowiak et al., 2020; Zhang et al., 2021). The pangenome of a species is composed of the core genome that is present in all individuals and the dispensable genome (PAV) that is present in some individuals of a species (Morgante et al., 2007; Marroni et al., 2014). Polyploid genomes may also be more prone to containing structural variation that could play an important role in phenotypic variance (Schiessl et al., 2018).
Wheat production in many countries, where rain tends to fall during the harvest season, is negatively impacted by pre‐harvest sprouting (PHS), the precocious germination of grains before harvest. PHS is directly associated with reductions in both grain yield and flour quality. It is estimated that PHS damage is responsible for up to $1 billion in annual losses (Nakamura, 2018; Vetch et al., 2019). In wheat, compared with red‐grained cultivars, white‐grained cultivars are characterized by more efficient flour extraction, a better ash content, a more favorable appearance and a less bitter taste in the final product (Ransom et al., 2006). However, white‐grained wheat has been reported to be more susceptible to PHS than red‐grained wheat for more than 100 yr (Nilsson‐Ehle, 1914; Gordon, 1979; Warner et al., 2000). The pigment of red‐grained wheat is presumed to be a result of the accumulation of the polyphenolic compound phlobaphene, which is derived from the precursor catechin and is an end product of the flavonoid pathway (Miyamoto and Everson, 1958; Gordon, 1979). The R2R3‐MYB transcription factor Myb10, located on wheat chromosomes 3A/3B/3D, encodes the R‐1 gene that controls redness by regulating the accumulation of anthocyanins (Groos et al., 2002; Himi & Noda, 2005; Himi et al., 2005; Himi et al., 2011). Myb10 is known to regulate flavones, flavonols, and proanthocyanidin by regulating many genes in the pathways of flavonoid biosynthesis (Lai et al., 2013). PHS resistance genes that span the chromosomal region of Myb10 have been discovered in red‐grained wheat (Groos et al., 2002; Rasul et al., 2009). However, the relationship between Myb10 and PHS resistance genes is still unclear (Medina‐Puche et al., 2014).
Common wheat is an allopolyploid species originating from hybridization between tetraploid Triticum turgidum (AABB) (McFadden & Sears, 1946) and Aegilops tauschii (DD) (Kihara, 1944) in the Fertile Crescent, where little rain falls during the harvest season, and PHS is therefore rare (Nakamura, 2018). Aegilops tauschii exhibits a high degree of PHS resistance (Liu et al., 1998). Hybrids have been recreated by crossing T. turgidum with A. tauschii to produce so‐called synthetic hexaploid wheat (SHW) to analyze PHS resistance (Imtiaz et al., 2008; Yang et al., 2014). PHS‐3D is an SHW‐derived locus linked to Myb10‐D (TaMyb10 on D genome of wheat) and confers strong PHS resistance explaining up to 42.47% of PHS variation in seven environments (Yang et al., 2019). Here, we demonstrate that PHS‐3D is Myb10‐D and negatively controls germination by activating 9‐cis‐epoxycarotenoid dioxygenase (NCED) transcription, which is involved in ABA biosynthesis; deepening grain color and increasing barriers to water uptake in wheat.
Materials and Methods
Plant materials
SHW‐L1 is a medium PHS resistant synthetic wheat that carries PHS‐3D (QPhs.sicau‐3D) obtained from allohexaploidization between the deep seed dormancy A. tauschii AS60 (Liu et al., 1998) and PHS‐resistant T. turgidum AS2255 (Zhang et al., 2004). Chuanmai32 (CM32) is a PHS susceptible cultivated wheat (Yu et al., 2014). SHW‐L1 and CM32 derived recombinant inbred lines (RILs) were evaluated for PHS resistance in nine independent environments (Yang et al., 2019). One resistant RIL (RIL71) and one susceptible RIL (RIL143) were selected as parents to develop heterogeneous inbred family (HIF) populations with a total of 7500 F3:4 HIF lines for fine mapping. We assayed F2:3 lines with 17 pairs of KASP markers (Supporting Information Table S1) and generated enough F3:4 seeds. Then F3:4 lines with homozygous fragments were selected and phenotyped. The fine mapping population panel was planted in Chongzhou (30°36ʹ27.02″N, 103°37ʹ4.10″E) from 2016 to 2018 (2016‐CZ, 2017‐CZ, and 2018‐CZ). Each line was single‐seed planted in two 1.5 m long rows, with 50 cm between rows and 10 cm between plants within rows.
A natural population of 250 wheat from all over the world, as well as 16 accessions of diploid A. tauschii (2n = 2x = 14, DD), and two accessions of tetraploid T. turgidum ssp. durum (2n = 4x = 28, AABB) were collected from the US Department of Agriculture (USDA) and Sichuan Agricultural University (SICAU) (Wang et al., 2013; Zhou et al., 2018), and their germination percentages (GPs) were assayed previously (Table S2) (Zhou et al., 2017). These accessions were used to analyze the relationship between sequence variations and PHS resistance in wheat germplasms.
Evaluation of germination percentages and grain color
The wheat grain is a caryopsis with a pericarp. This article refers to the wheat caryopsis as a ‘seed’ for simplicity’s sake. Both whole spikes and threshed seeds were used for the germination tests. The materials were harvested when the grains reached physiological maturity and then air‐dried in the shade at room temperature until seed moisture was < 10%. Three replications of 50 threshed undamaged seeds were incubated in Petri dishes that contained filter paper in the lower half and 5 ml of distilled water at 20°C without light for 7 d. The GPs (i.e. total germinated seeds/total assayed seeds) was used to assay the germination level at 7 d after imbibition (DAI) (Walker‐Simmons, 1988). For each independent plant, only mature grains from primary or secondary spikes were selected to be stored at −20°C until phenotyping. In total, 20 spikes collected from 16 plants were wrapped in moist filter paper after being immersed in distilled water for 6 h, and put in a dew chamber without light at 20°C for 7 d and photographs taken every day. Seeds in spikes were threshed and the GP was calculated on the seventh day (Paterson & Sorrells, 1990).
The seeds were scanned with an Uniscan M2800 Scanner (Unis, China) Series_V2.0, and the scanned images were analyzed by Photoshop CS6 straw tool (Stetter et al., 2020). Seed colors were inferred visually from seed samples.
Evaluation of seed coat permeability
Seed coat permeability is important to study as it plays significant roles in seed dormancy and germination, thus the seed permeability was checked with the TTC (2,3,5‐triphenyltetrazolium chloride) method (Roistacher et al., 1957). During seed imbibition, higher permeability could be indicated by the deeper dyeing color of TTC. Germinating seeds of transgenic lines (overexpression, OE) and control (wild‐type, WT) were added by 1% TTC and incubated at 37°C. The color variance of seeds was artificially assayed at 3, 6, 12, 24, 36, 48 and 60 h after imbibition (HAI), and used to determine seed coat permeability.
Genomic sequences data
The genomic reference data of Chinese Spring (ftp://ftp.ensemblgenomes.org/pub/plants/current/fasta/triticum_aestivum/dna/), AL8/78 (http://aegilops.wheat.ucdavis.edu/ATGSP/), Cadenza, and Robigous (https://opendata.earlham.ac.uk/opendata/data/Triticum_aestivum/EI/v1.1/) were obtained from the database. And the data of Aikang58 were obtained from Dr Jizeng Jia (Jia et al., 2021) (Table S3). The genotyping data of 12 hexaploid wheat and one tetraploid wheat cultivars from the wheat pan‐genome database (http://www.10wheatgenomes.com/) were also analyzed (Walkowiak et al., 2020).
Additional materials and methods
Details on the DNA extraction and genotyping (Methods S1), fine mapping of PHS‐3D (QPHS.sicau‐3D) (Methods S2), optical map construction (Methods S3), gene cloning and sequencing (Methods S4), RNA preparation and RNA sequencing (Methods S5), next‐generation sequencing (NGS) data analysis (reads mapping, assembly, and functional annotation) (Methods S6), analysis of gene expression by quantitative polymerase chain reaction (qPCR) (Methods S7), gene transformation (Methods S8), endogenous ABA profiling (Methods S9), flavonoids extraction and profiling (multiple reaction monitoring, MRM) (Methods S10), subcellular localization of Myb10‐D (Methods S11), RNA in situ hybridization (Methods S12), protein expression in Escherichia coli and purification (Methods S13), electrophoretic mobility shift assay (EMSA) (Methods S14), transient expression assay (Methods S15), yeast one‐hybrid (Methods S16), statistical analyses (Methods S17), and code used in this study (Methods S18) can be found in the Supporting Information.
Results
PHS‐3D is located within a large PAV region
PHS‐3D (QPHS.sicau‐3D) was previously detected in a RIL population between the synthetic wheat genotype SHW‐L1 and common wheat cultivar Chuanmai 32 (CM32) (Yang et al., 2014). Here, by mapping SNP markers in the region of PHS‐3D (Fig. 1a), two RILs, i.e. the PHS resistant line L71 and susceptible line L143, were selected to develop HIF populations (Fig. 1b). A total of 7500 F3:4 families that reckoned to the population comprising self‐progenies of selected individuals with heterozygous PHS‐3D from 50 F2:3 plants. Then, we developed a total of 17 markers to fine‐map PHS‐3D using the Kompetitive Allele‐Specific PCR (KASP) technique based on SNP in this region (Supporting Information Fig. S1; Table S1). By combining the genotype and phenotype, the PHS‐3D region was narrowed to a 2.7 Mb region, flanking from K‐AX‐108730423 (572.2 Mb) to K‐AX‐111501911 (574.9 Mb). Though 12 out of 17 KASP markers were located in this 2.7 Mb region, their signal could not be detected in 836 PHSS F3:4 lines. Thus, no further recombination was observed, which indicated that there was a lack of recombination (Fig. 1c).
Fig. 1.
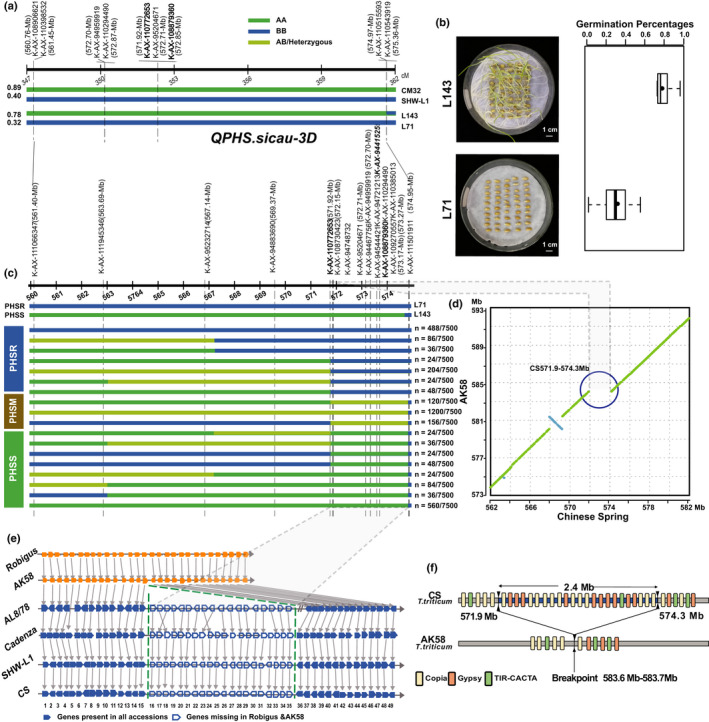
Fine mapping and genomic analysis of PHS‐3D from wheat (Triticeae aestivum). (a) QTL of the QPHS.sicau‐3D (PHS‐3D) region in pre‐harvest sprouting (PHS)‐resistant SHW‐L1, PHS‐susceptible CM32, and their recombinant inbred lines (RILs) L143 and L71. (b) The average germination percentage of L143 (77.1%) and L71 (29.6%) from six environments at 7 d after imbibition (DAI). (c) Fine mapping of PHS‐3D. PHSR, PHSS, and PHSM indicated PHS resistance, susceptibility, and moderate resistance, respectively. The numbers on the right of the bar indicate the number of recombinants. The two markers (i.e. AX‐108730423 and AX‐108925806) were close to PHS‐3D, and two markers (i.e. AX‐110772653 and AX‐108879360) were identified in our previous genome‐wide association study (GWAS) and bi‐parental QTL mapping analysis. The physical positions of 17 KASP markers are based on the IWGSC CS RefSeq v2.0 assembly. (d) The dot‐plot shows a 2.4 Mb presence–absence variation (PAV) between Chinese Spring (CS) and Aikang58 (AK58). (e) Twenty annotated genes (gene order: 16‐35) are missing in the PAV region. (f) Transposable elements, including LTR_copia, LTR_gypsy, and Tir_CACTA, are abundant in the PAV region. SD, error bars given in the graphs.
The absence of recombination is related to a PAV. A total of 1534 Gb of large single molecules (> 150 Kb) labeled by the DLE‐1 enzyme was obtained from the PHS‐susceptible cultivar CM32, and de novo assembled into a BioNano optical map. Taking advantage of the optical map, we identified a deletion (Chr3D: 571.5–574.4 Mb) in CM32 by aligning its genome sequence to the Chinese Spring (CS) genome sequences. Further sequence comparison revealed an ~2.4 Mb deletion in the common wheat cultivars Robigus, and Aikang58 (AK58, Chr3D: 583.6–583.7 Mb) (Fig. 1d) relative to CS (Chr3D: 571.9–574.3 Mb), Cadenza, and the synthetic wheat genotype SHW‐L1 (Fig. 1e; Table S3). The deleted region resulted in the loss of 20 genes/pseudogenes (gene order: 16–35), whereas the genes flanking the PAV region (gene order: 1–15 and 36–49) were conserved (Table S4). The PAV region harbored 23 transposable elements (LTR/Copia, Gypsy, and TIR) in CS (Fig. 1f). Based on the molecular signatures at the breakpoints, unequal crossing over between TEs may have led to the PAV in wheat (Table S5).
The large PAV is associated with PHS resistance in wheat germplasms
We developed 14 sequence‐tagged‐site (STS) markers for genes/pseudogenes in the PAV region (Fig. S2; Tables S6) and carried out an association analysis using 16 A. tauschii (DD genome), three T. turgidum (AABB genome), and 262 hexaploid wheat (AABBDD genome) lines to assess the relationship between PAV and PHS resistance (Fig. 2a). As expected, no PCR products were detected in T. turgidum due to the absence of the D genome. A total of three haplotypes were identified among the wheat lines, i.e. Hap‐I without a deletion, Hap‐II with a 2.2 Mb deletion, and Hap‐III with the whole 2.4 Mb deletion (Fig. 2b). Hap‐I and Hap‐III are more prevalent than Hap‐II in common wheat. This 2.4 Mb region in common wheat is also conserved in 16 analyzed A. tauschii accessions collected from Caspian Sea areas.
Fig. 2.
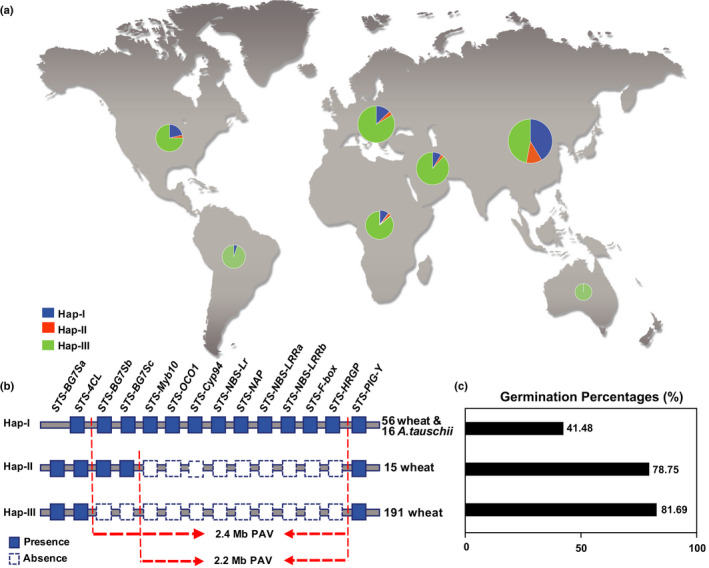
Presence–absence variation (PAV) of PHS‐3D in wheat (Triticeae aestivum) germplasms. (a) Distribution of three haplotypes (Hap) characterized in 262 wheat and 16 Aegilops tauschii. (b) Constitution of the three haplotypes. (c) Average germination percentages of the three haplotypes.
The average GPs of the wheat lines of Hap‐I (GP: 41.48%) were significantly lower than those of Hap‐II (GP: 78.75%, u = 8.55) and Hap‐III (GP: 81.69%, u = 9.85) (Fig. 2c; Table S2). In Hap‐I, 98.44% of lines were predominately red (brown, tan)‐grained, and only one landrace AS661609 (Tuotuomai), collected from China, was white‐grained. The reduction in PHS resistance in Hap‐II and Hap‐III indicated that PHS‐3D is probably located in the deleted region between STS‐Myb10 (572.2 Mb) and STS‐HRGP (573.2 Mb).
Myb10‐D is highly expressed in the seeds of PHS resistant wheat
The 20 genes located within the 2.4 Mb deleted region that probably harbors PHS‐3D were used to identify the functional gene(s) that contribute to PHS resistance. Transcriptomic results obtained for seeds 12 and 24 HAI, seedling leaves, roots, and seeds 10, 20 and 30 d post‐anthesis (DPA) were compared between SHW‐L1 and CM32 (Fig. S3a; Table S6). All of the 20 analyzed genes were not expressed in CM32 at a level of transcripts per million (TPM) > 1, except RS19 (gene #33, chloroplastic 30S ribosomal protein S19, which had 92 copies in the wheat genome). Three genes Myb10, TAF9, and Oco1 were expressed in the seed of SHW‐L1. TAF9 is an orthologous gene for TATA‐binding protein‐associated factor 9 that is associated with salt and heat stress responses (Li et al., 2017). Oco1 is a homolog of oral cancer‐overexpressed protein 1 (ORAOV1) (Jiang et al., 2010). Myb10‐D is known as an R2R3‐MYB transcription factor. Then we checked their expression levels in 29 tissues/stages of CS and BCScv1 (Fig. S3b) (IWGSC et al., 2018). TAF9 and OCO1 are highly expressed in all tissues (root, shoot, leaf, flower, and seed), with 18 and six orthologs in common wheat, respectively. Myb10 is highly expressed in the seed (2DPA) and aleurone layer (12DPA). At last, we confirmed the high expression of Myb10 in SHW‐L1 at 5‐15 DPA by qPCR (Fig. S3c). Thus, Myb10‐D was prioritized for functional characterization because it has been reported as a QTL controlling PHS resistance (Himi & Noda, 2005).
The PHS‐susceptible soft white spring wheat cultivar Fielder (WT) without the 2.4 Mb region (Table S2) was transformed with Myb10‐D driven by the polyubiquitin promoter (Ubi1). Then, we assessed the Myb10‐D expression pattern by RNA in situ hybridization (Fig. S4). The signals indicated that the expression of Myb10‐D was concentrated mainly in the mesocarp of the inner pericarp in the positive lines 5–15 DPA. This result was consistent with the observation that Myb10‐D was predominantly expressed in wheat seeds.
PHS resistance is enhanced in Myb10‐D overexpressed transgenic wheat
We found that seeds of the OE lines were red‐grained, while seeds of the WT control were white‐grained (Fig. 3a). Compared to that of the WT, the germination of seeds from the OE lines was significantly delayed for both whole spike and threshed seeds (Table S7; Fig. 3b). Myb10‐D led to delayed germination (Fig. 3c) and decreased the GP of 55.1% in threshed seeds and 38.5% in whole spikes (Fig. 3d). The intensity of TTC red coloration in WT seeds was higher than that in seeds from the OE lines and was proportional to seed coat permeability (Fig. 3e). These results support the assumption that the Myb10‐D identified as PHS‐3D is involved in PHS resistance in wheat.
Fig. 3.
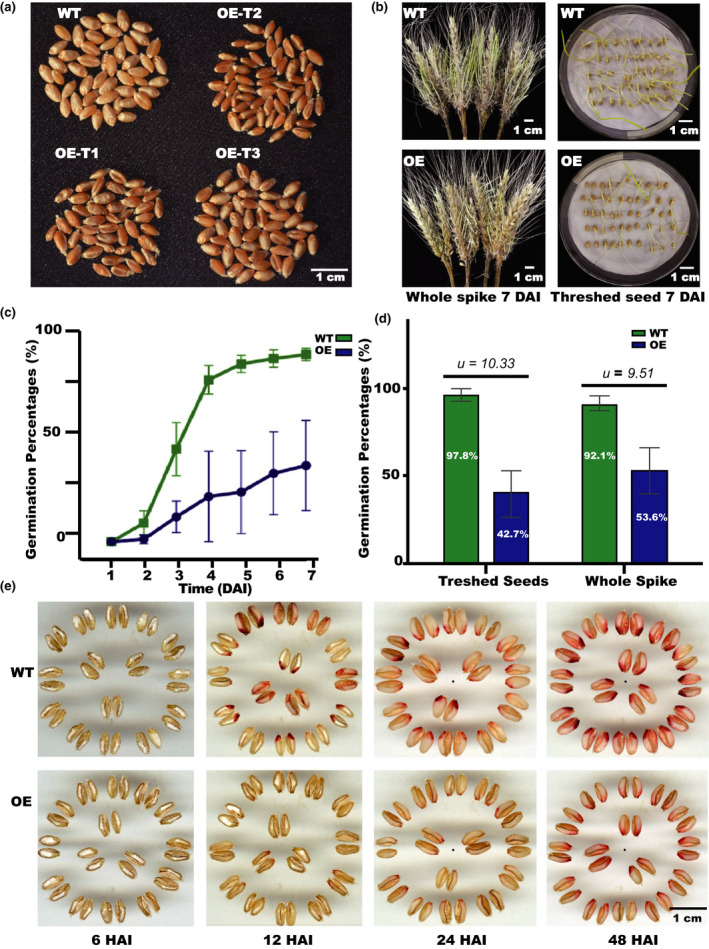
Characterization of Myb10‐D functions in transgenic plants. Phenotypic variation between wild‐type (WT) and Myb10‐D overexpression (OE) wheat (Triticeae aestivum) lines, including the germination percentages (GPs) of the whole spikes and thrashed seeds, as well as their seed coat permeability. (a) Profile of seeds in the T1–T3 generations. (b) GP in whole spikes and threshed seeds of OE lines (T3 generation) and WT plants. (c) GP of threshed seeds of OE and WT lines 1–7 d after imbibition (DAI). (d) The GPs were 55.1% and 38.5% lower in the OE lines than in the WT for threshed seeds and whole spike, respectively. (e) Seed coat permeability assumed to be associated with color was assayed by eye. Note: u > 2.58 (99% confidence interval); HAI, hours after imbibition. SD, error bars given in the graphs.
Myb10‐D regulated the biosynthesis of flavonoids and ABA
Subcellular localization showed that the fused protein 35S::Myb10‐D:GFP was located in the nucleus of Nicotiana benthamiana leaf protoplasts (Fig. S5). Thus, the nuclear localization of Myb10‐D was expected as a transcription factor that is a nuclear protein and may regulate genes during seed development.
The integrated transcriptomic and metabolomic analysis revealed changes in the concentrations of both flavonoids and abscisic acid (ABA) in OE lines compared with WT lines. To uncover the variance in grain color and germination between WT and OE lines, we assayed the expression of genes related to flavonoid and ABA biosynthesis and the accumulation of these compounds by RNA‐sequencing (RNA‐seq), high‐performance liquid chromatography (HPLC), and electrospray ionization triple quadrupole linear ion‐trap tandem mass spectrometry (ESI‐QTRAP‐MS/MS) (Fig. 4). The expression levels (TPM) of Myb10‐D were 63.19 and 34.21 in the OE lines at 5 DPA and 10 DPA, respectively, while no expression was detected in the WT (Table S8). Genes involved in the methyl‐erythritol phosphate (MEP) and flavonoid pathways, which determine the biosynthesis of flavonoids and ABA (Shirley et al., 1995; Milborrow, 2001), exhibited positive correlations between metabolite and transcript accumulation in the OE lines. Myb10 was previously described as the R‐1 gene, a seed‐specific gene that regulates flavonoid metabolites by promoting the expression of genes (PAL, FLS, CHS, CHI, F3H, GT, DFR, ANS, F3′H and UFGT) in the biosynthetic pathways (Himi & Noda, 2005; Lai et al., 2013). In this study, eight genes in the flavonoid biosynthesis pathway (CHS, CHI, F3H, GT, DFR, ANS, F3′H and UFGT) were upregulated in the OE lines (Table S9). Four genes in the MEP biosynthesis pathways (AAO, NCED, ABA3 and SDR) with upregulated expression patterns similar to that of Myb10‐D were detected. The accumulation of ABA increased one‐fold in the seeds from OE compared to WT. And the deeper grain color in the OE lines resulted from the significantly higher biosynthesis of flavonoid 3‐glucosyl isorhamnetin, tricin, salcolin B, nicotiflorin, narcissin, quercetin, rutin, and cynaroside (Fig. 3a). The observed increases in both ABA and flavonoids, as well as the upregulation of associated genes, in Myb10‐D OE lines, indicated that Myb10‐D also regulates genes in the ABA biosynthetic pathway. The genes associated with flavonoids have been well studied. Thus, we selected AAO, NCED, ABA3 and SDR for a more in‐depth examination.
Fig. 4.
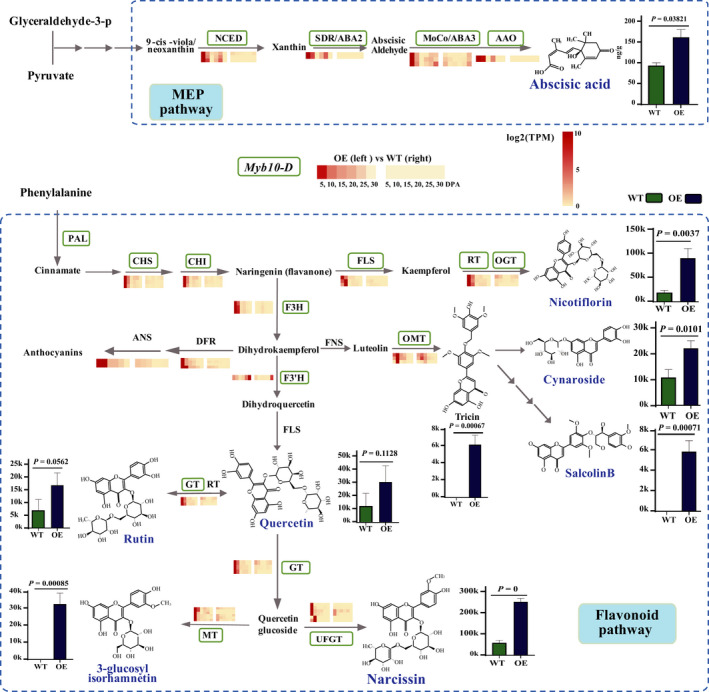
Myb10‐D regulates the biosynthesis of flavonoids and abscisic acid (ABA) via the methyl‐erythritol phosphate (MEP) and flavonoid pathways in wheat (Triticeae aestivum). The details of expression for ABA2, ABA3, AAO and NCED in the MEP pathway, and CHS, CHI, F3H, F3’H, ANS, DFR, GT, MT and UFGT in flavonoid pathway are provided in Supporting information Table S8. The accumulations of flavonoid 3‐glucosyl isorhamnetin (> 100×), tricin (> 100×), salcolin B (> 100×), nicotiflorin (4.68×), narcissin (4.28×), quercetin (2.45×), rutin (2.41×), cynaroside (2.04×), and ABA (2×) increased (Table S9). SD, error bars given in the graphs.
Myb10‐D controls germination by positively regulating NCED transcription
The expression levels were confirmed by qPCR except for AAO whose expression was similar to WT (Figs 5a, S6a). Then 2 kb promoter region of NCED, ABA3, and SDR was found to carry cis‐elements highly similar to the consensus Myb binding secondary wall MYB‐responsive element (SMRE) of ACC(A/T)A(A/C)(T/C) and ACC(A/T)ACC(A/C/T) (Grotewold et al., 1994; Zhong & Ye, 2012) (Fig. 5b). To verify possible binding, we carried out an EMSA (Fig. 5c). The Pcold‐Myb10‐D fusion protein could bind to fluorescently labeled DNA probes containing binding motifs but failed to bind to mutant probes. This binding was not detected under incubated with His protein (without Myb10‐D protein). To further examine the specificity of the binding, WT competitors and mutated competitors were introduced. The binding affinities of ABA3 and NCED gradually decreased with increasing concentrations of WT competitor probes but were not affected by mutated competitor probes (Figs 5c, S6b). Adding Myb10‐D protein caused reduced gel migration of ABA3 and NCED, indicating the formation of a DNA–protein complex. However, no significant difference was detected for AAO or SDR.
Fig. 5.
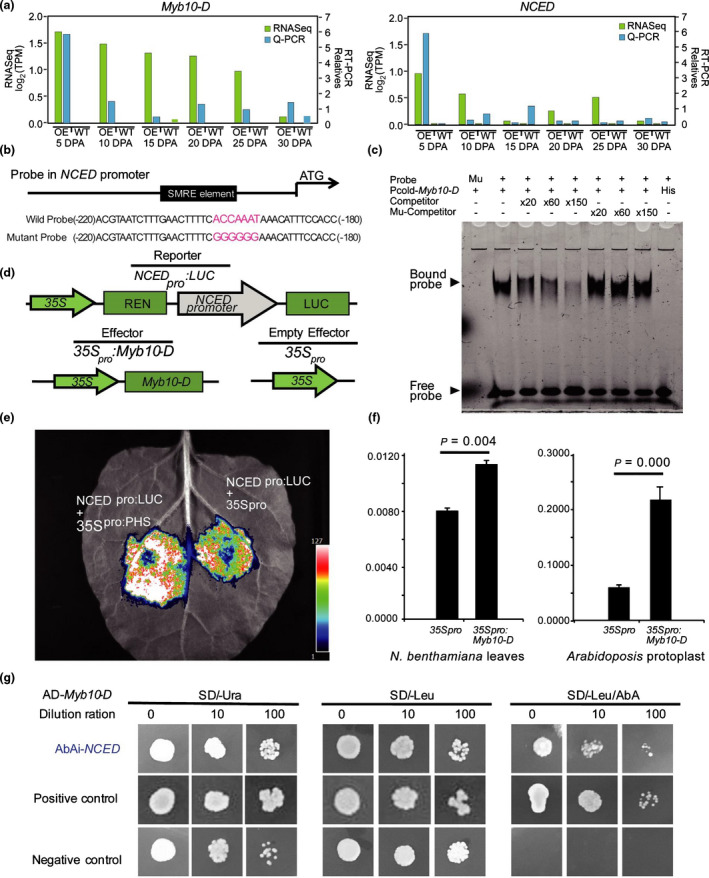
Myb10‐D binds to the secondary wall MYB‐responsive element (SMRE) in the NCED promoter and induces its expression in wheat (Triticeae aestivum). (a) The expression profiles of Myb10‐D (left) and NCED (right) 5‐30 DPA as detected by RNA‐seq and qPCR. (b) Schematic diagram of the 9‐cis‐epoxycarotenoid dioxygenase (NCED) promoter structure. The SMRE element is −192 to −199 bp away from the ATG of NCED (TraesCS6B02G298800). (c) Electrophoretic mobility shift assay (EMSA) of Mb10‐D with probes containing the binding motifs in the promoters of NCED. The presence (+) or absence (–) of specific probes is indicated. (d) Schematic diagram of various effector and reporter constructs of NCED. (e) Dual‐luciferase assay of Myb10‐D in the promoters of NCED in Nicotiana benthamiana leaves. (f) Significant differences (Student's t‐test: *, P < 0.05) detected in N. benthamiana leaves and Arabidopsis protoplasts. Firefly luciferase (LUC) activity was normalized to the Renilla luciferase (REN) activity (as an internal control). Values are means ± SD from three independent replicates for each construct. (g) Yeast one‐hybrid analysis of the interaction of Myb10‐D and the NCED promoter. The promoter was used for autoactivation tests in the presence of different concentrations of aureobasidin A (AbA) on SD/‐Ura medium, and physical interaction was determined on SD/‐Leu medium in the presence of corresponding AbA concentrations.
We used a dual‐luciferase reporter (DLR) system to identify how Myb10‐D affects the expression of ABA3 (TraesCS7A02G269200) and NCED (TraesCS6B02G298800 with 98% identity to TaNCED, GenBank: KP0900105). Coexpression of a construct containing the Myb10‐D full‐length coding sequence with the NCEDpro:LUC reporter (Fig. 5d) led to significantly higher luciferase (LUC) expression in 35Spro:Myb10‐D than in the 35Spro control from both N. benthamiana leaves (P = 0.004) (Fig. 5e) and Arabidopsis protoplasts (P = 0.000) (Fig. 5f), but no significant difference was detected for ABA3pro:LUC (Fig. S6c). This result corroborates that Myb10‐D directly binds to the SMRE at −192 to −199 bp in the NCED promoter and functionally activates its expression.
The interaction between Myb10‐D and the NCED promoter was confirmed by a yeast one‐hybrid assay (Fig. 5g). The promoter of NCED was inserted into the pAbAi vector (Clontech, Japan) and transformed into the Y1H strain. The NCED promoter was not activated without protein binding. Yeast containing Myb10‐D exhibited normal growth under 400 ng ml−1 aureobasidin A (AbA). In contrast, the growth of the negative control cells containing the empty vector was inhibited, which indicated that Myb10‐D could directly bind to the promoter of NCED. Collectively, these results support that Myb10‐D promotes ABA synthesis, repressing germination by positively regulating NCED transcription.
Discussion
We have demonstrated that Myb10‐D determines PHS resistance by using five complementary approaches: (1) fine mapping of PHS‐3D that explains up to 42.5% of PHS variation in a 2.4 Mb PAV region; (2) an assessment of the variation of PHS‐3D and its association with PHS resistance (about 40% lower GP with PHS‐3D than those without it in globally‐sourced wheat germplasms); (3) detection of the mechanism by which Myb10‐D causes the PHS‐3D effect by integrated genomic and transcriptomic analyses; (4) phenotyping of plant metabolomic phytohormones, transcriptomes, and germination in transgenic lines (38.5–55.1% lower GP in OE than that in WT lines); and (5) characterization of the role of Myb10‐D in promoting NCED in ABA biosynthesis.
PHS‐3D, located in a PAV region, was hard to characterize by regular fine mapping and map‐based cloning strategies until recent developments in the omics technology of wheat. Plant genome structural variations, including copy number variations, presence and absence variations, inversions, and translocations, can be beneficial, neutral, or deleterious to the plant (Saxena et al., 2014). In the wheat pangenome project, there were 81070 ± 1631 genes and an average of 128 656 genes in each of 18 elite cultivars compared with CS, and only 2/3 genes shared by all cultivars (Montenegro et al., 2017). Here, we uncover a new case in which the important agronomic traits grain color and PHS among wheat lines are affected by PAV. We found that the dual function Myb10‐D is in a 2.4 Mb PAV region on 3DL, where unequal crossing over occurred among repetitive LTR TEs with high sequence similarity at the breakpoints (Fig. 1). Because of this large PAV, analyzing the relationship between grain color and germination is difficult without further recombinational events in bi‐parental linkage mapping studies. This is the reason that we have known about the relationship between grain color and germination for more than 100 yr, but it has been challenging to uncover the mechanism and to breed white‐grained PHS resistant cultivars. Our results indicate the strategy of the white grained PHS resistant wheat cultivar breeding using white grained wheat with Myb10‐D. However, only one white‐grained wheat landrace (Tuotuomai/Totoumai) with Myb10‐D is PHS resistant (Table S2). We will check the variation of the downstream genes associated with the flavonoids pathway in the white dormancy landrace Tuotuomai in the future. This kind of germplasm will be a useful resource for white‐grained wheat breeding (Chen et al., 2008; Zhu et al., 2014; Zhou et al., 2017).
Myb10‐D was significantly (P < 0.001) associated with the PHS resistance in wheat varieties and advanced lines (Wang et al., 2016). However, it has been reported that red grain color and seed dormancy can be separated by recombination (DePauw & McCaig, 1983). The ability to separate these traits indicates that grain color and dormancy may be only genetically linked instead of controlled by the same gene (Warner et al., 2000). We observed that in white‐grained wheat, in which the 2.4 Mb region on 3DL is absent, the grain is both lighter in color and germinates faster (Figs 1, 2). White‐grained wheat is homozygous for recessive Myb10‐D, which could lead to both PHS resistance and the red grain phenotype from red‐grained wheat. Although the Myb10‐D in wheat and its D genome donor A. tauschii are conserved, it is not present in 22 landraces from Turkey, Iran, India, Nepal, Iraq, Pakistan, and Jordan, indicating that the deletion could have occurred in the early stages of wheat cultivation. Relatives of wild wheat—such as A. tauschii—are more dormant (conferring PHS resistance) than domesticated wheat, indicating that artificial selection has contributed to the reduction of dormancy in modern wheat (Dong et al., 2015; Volis, 2016; Zhang et al., 2017). Likewise, white rice appears to show the domestication syndrome and remains under strong selection in most rice breeding programs (Sweeney et al., 2006). The fact that white‐grained wheat is ubiquitous suggests that the deletion also drove the critical transition from the deep seed dormancy and asynchronous germination observed in A. tauschii to the shallow and uniform germination observed in wheat cultivars. However, it also resulted in PHS susceptibility.
Our results indicate that Myb10‐D regulates the transcription levels of genes involved in the MEP and flavonoid pathways, which regulate the biosynthesis of ABA and flavonoids, respectively. Here, the synthesis of ABA and flavonoids was increased, and seed coat permeability was decreased in the OE lines (Figs 3, 4). It has been shown that the presence of phenolic flavonoids influences water uptake and may also hinder imbibition, while barriers to water uptake owing to pigmentation may influence dormancy (Huang et al., 1983; Rathien et al., 2009). NCED cleaves 9‐cis‐violaxanthin and 9′‐cisneoxanthin to produce xanthoxin as a precursor of ABA and delay wheat seed germination (Kende & Zeevaart, 1997; Tong et al., 2016). In addition to regulating flavonoids, we found that Myb10 involves the ABA synthesis by promoting NCED. Moreover, we will check the ABA levels at more time points during seed development and after‐ripening by time‐course experiments. We propose a hypothetical working model for the Myb10‐D dependent regulation of delayed seed germination (Fig. 6). In this model, Myb10‐D regulates the expression of genes in the flavonoid pathway and regulates NCED by binding the SMRE in its promoter region, ultimately affecting the concentrations of flavonoids and ABA. Thus, the dual role Myb10‐D can regulate both the grain color and seed germination of wheat.
Fig. 6.
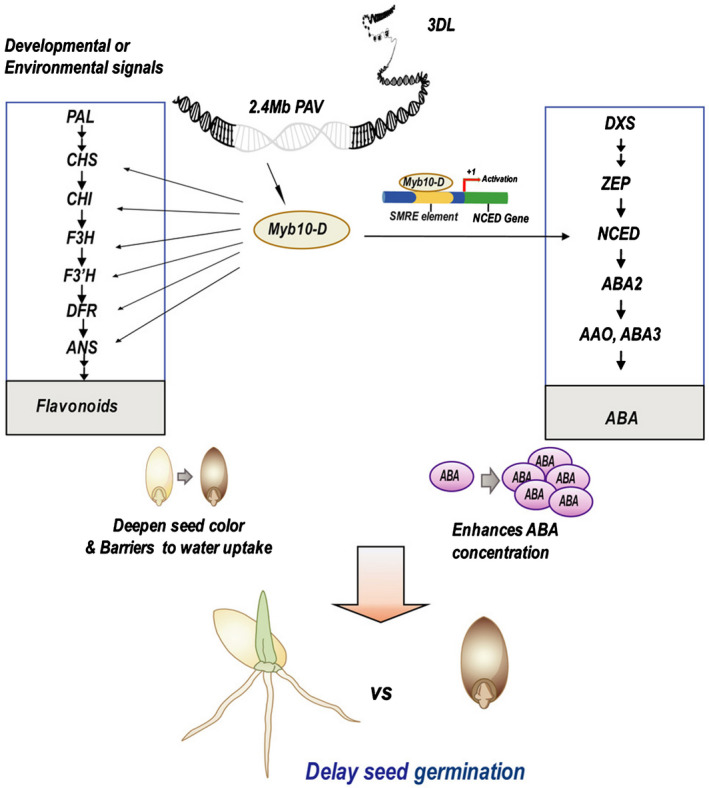
Hypothetical working model of Myb10‐D dependent regulation of grain color and germination mediated by flavonoids and abscisic acid (ABA) in wheat (Triticeae aestivum). PAV, presence–absence variation; SMRE, secondary wall MYB‐responsive element.
Author contributions
Plant materials: MH, DL, YZheng, YWei, PQ, ZP, JM, QJ, GC, LL, LH. Comparative genetics and RNA‐seq of candidate genes: MC, JRW, XG, LG, MD, ZC. Divergence of the resistant gene in germplasms: MD, JRW. Experiment design and data analysis: JRW, YWu, M‐CL, DL. Fine mapping and map‐based clone: JL, JRW, YZhou, LZ, JY, MH, TZ, YY. Gene functional analysis: YF, JL, ML, YL. Manuscript writing: JRW, YZhou, MC, JL, DL, YF, YWu. Phenotype analysis: JL, JRW, ML, ZL, CT. JL, YF and YZhou contributed equally to this work.
Supporting information
Fig. S1 KASP markers were developed by SNPs in the QPHS.sicau‐3D (PHS‐3D) region.
Fig. S2 STS markers were developed in the 2.4 Mb PAG region on 3DL for genotyping wheat lines.
Fig. S3 Expression profiles of genes in the PAV region containing PHS‐3D.
Fig. S4 RNA in situ hybridization of Myb10‐D in transversal sections 5‐15 DPA indicates its expression in the mesocarp of the inner pericarp.
Fig. S5 Subcellular localization of Myb10‐D in the nucleus.
Fig. S6 Characterization of binding sites based on the 2‐kb promoter regions of potential Myb10‐D regulated genes in the MEP pathway.
Methods S1 DNA extraction and genotyping.
Methods S2 Fine mapping of PHS‐3D (QPHS.sicau‐3D).
Methods S3 Optical map construction.
Methods S4 Gene cloning and sequencing.
Methods S5 RNA preparation and RNA sequencing.
Methods S6 NGS data analysis (reads mapping, assembly, and functional annotation).
Methods S7 Quantitative PCR with reverse transcription (RT)‐PCR.
Methods S8 Gene transformation.
Methods S9 Endogenous ABA profiling.
Methods S10 Flavonoids extraction and profiling (multiple reaction monitoring, MRM).
Methods S11 Subcellular localization of Myb10‐D.
Methods S12 RNA in situ hybridization.
Methods S13 Protein expression in Escherichia coli and purification.
Methods S14 Electrophoretic mobility shift assay.
Methods S15 Transient expression assay.
Methods S16 One‐hybrid assays in yeast.
Methods S17 Statistical analyses.
Methods S18 Code used in this study.
Table S1 Primers used for KASP, STS, PCR clone, qPCR, transgenetic, and transactivation assay.
Table S2 Genotyping and phenotyping of 262 common wheat, 16 Aegilops tauschii, and three tetraploid wheat lines.
Table S3 Summary of genomic reference sequences data and NGS data obtained in this study (NCBI, PRJNA605937).
Table S4 Genes in the 2.4 Mb PAV region from six accessions CS, AK58, AL8/78, Robigus, Cadenza, and SHW‐L1.
Table S5 Transposons in the 2.4 Mb PAV region from CS and AK58.
Table S6 The expression of 20 genes in 2.4 Mb PAV region from CM32 and SHW‐L1 in 10‐30 DPA seeds, and the expression of three selected genes from CS and BCScv1 in 29 tissues or stages.
Table S7 Germination percentages of WT and OE lines.
Table S8 Different expression genes that had similar expression patterns with Myb10‐D form WT and OE lines in 5‐30 DPA seeds.
Table S9 Accumulation of flavonoids and ABA in WT and OE lines.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
The authors thank Dr Jizeng Jia from the Chinese Academy of Agricultural Science to provide the data of Aikang58. This work was supported by the National Key Research and Development Program of China (2018YFE0112000; 2017YFD0100900), the National Natural Science Foundation of China (31871609, 91935303, 31571654 and 31171555), the National Basic Research Program of China (2014CB147200), and the Sichuan Science and Technology Support Project (2021YFH0077).
Data availability
The sequence data that support the findings of this study are openly available in NCBI Sequence Read Archive at https://www.ncbi.nlm.nih.gov/, under accession number PRJNA605937. Other main data that supports the findings of this study are available in the Supporting Information of this article.
References
- Chen CX, Cai SB, Bai GH. 2008. A major QTL controlling seed dormancy and pre‐harvest sprouting resistance on chromosome 4A in a Chinese wheat landrace. Molecular Breeding 21: 351–358. [Google Scholar]
- DePauw RM, McCaig TN. 1983. Recombining dormancy and white seed color in a spring wheat cross. Canadian Journal of Plant Science 63: 581–589. [Google Scholar]
- Dong ZD, Chen J, Li T, Chen F, Cui DQ. 2015. Molecular survey of Tamyb10‐1 genes and their association with grain colour and germinability in Chinese wheat and Aegilops tauschii . Journal of Genetics 94: 453–459. [DOI] [PubMed] [Google Scholar]
- Gordon IL. 1979. Selection against sprouting damage in wheta: III. Dormancy, germinative alpha‐amylase, grain redness, and flavanols. Australian Journal of Agricultural Research 30: 387–402. [Google Scholar]
- Groos C, Gay G, Perretant MR, Gervais L, Bernard M, Dedryver F, Charmet G. 2002. Study of the relationship between pre‐harvest sprouting and grain color by quantitative trait loci analysis in a white × red grain bread‐wheat cross. Theoretical and Applied Genetics 104: 39–47. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Drunnond BJ, Bowen B, Peterson T. 1994. The myb‐homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553. [DOI] [PubMed] [Google Scholar]
- Himi E, Maekawa M, Miura H, Noda K. 2011. Development of PCR markers for Tamyb10 related to R‐1, red grain color gene in wheat. Theoretical and Applied Genetics 122: 1561–1576. [DOI] [PubMed] [Google Scholar]
- Himi E, Nisar A, Noda K. 2005. Colour genes (R and Rc) for grain and coleoptile upregulate flavonoid biosynthesis genes in wheat. Genome 48: 747–754. [DOI] [PubMed] [Google Scholar]
- Himi E, Noda K. 2005. Red grain color gene (R) of wheat is a Myb type transcription factor. Euphytica 143: 239–242. [Google Scholar]
- Huang G, McCrate AJ, Varriano‐Marston E, Paulsen GM. 1983. Caryopsis structural and imbibitional characteristics of some hard red and white wheats. Cereal Chemistry 60: 161–165. [Google Scholar]
- Imtiaz M, Ogbonnaya FC, Oman J, van Ginkel M. 2008. Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross‐derived wheat lines. Genetics 178: 1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC) , Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A et al. 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: 7191. [DOI] [PubMed] [Google Scholar]
- Jia J, Xie Y, Cheng J, Kong C, Wang M, Gao L, Zhao F, Guo J, Wang K, Li G et al. 2021. Homology‐mediated inter‐chromosomal interactions in hexaploid wheat lead to specific subgenome territories following polyploidization and introgression. Genome Biology 22: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M, Guan J, Zhai Z, Geng S, Zhang X, Mao L, Li A. 2018. Wheat functional genomics in the era of next generation sequencing: an update. Crop Journal 6: 7–14. [Google Scholar]
- Jiang L, Zeng X, Wang Z, Ji N, Zhou Y, Liu XT, Chen QM. 2010. Oral cancer overexpressed 1 (ORAOV1) regulates cell cycle and apoptosis in cervical cancer HeLa cells. Molecular Cancer 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, Zeevaart J. 1997. The five "classical" plant hormones. Plant Cell 9: 1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara H. 1944. Discovery of the DD‐analyser, one of the ancestors of Triticum vulgare (Japanese). Agriculture & Horticulture (Tokyo) 19: 13–14. [Google Scholar]
- Lai Y, Li H, Yamagishi MA. 2013. A review of target gene specificity of flavonoid R2R3‐MYB transcription factors and a discussion of factors contributing to the target gene selectivity. Frontiers of Biology 8: 577–598. [Google Scholar]
- Li J, Yang J, Liu D, Huang R, Sui S, Li M, Zhang Q. 2017. Isolation and characterization of plant TAF9, an orthologous gene for TATA‐binding protein‐associated factor 9, from wintersweet (Chimonanthus praecox). Canadian Journal of Plant Science 97: 1100–1108. [Google Scholar]
- Liu DC, Lan XJ, Wang ZR, Zheng YL, Zhou YH, Yang JL, Yen C. 1998. Evaluation of Aegilops tauschii Cosson for preharvest sprouting tolerance. Genetic Resources and Crop Evolution 45: 495–498. [Google Scholar]
- Marroni F, Pinosio S, Morgante M. 2014. Structural variation and genome complexity: is dispensable really dispensable? Current Opinion in Plant Biology 18: 31–36. [DOI] [PubMed] [Google Scholar]
- McFadden ES, Sears ER. 1946. The origin of Triticum spelta and its free‐threshing hexaploid relatives. Journal of Heredity 37: 81–89. [DOI] [PubMed] [Google Scholar]
- Medina‐Puche L, Cumplido‐Laso G, Amil‐Ruiz F, Hoffmann T, Ring L, Rodriguez‐France A, Caballero JL, Schwab W, Munoz‐Blanco J, Blanco‐Portales R. 2014. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. Journal of Experimental Botany 65: 401–417. [DOI] [PubMed] [Google Scholar]
- Milborrow BV. 2001. The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. Journal of Experimental Botany 52: 1145–1164. [PubMed] [Google Scholar]
- Miyamoto T, Everson EH. 1958. Biochemical and physiological studies of wheat seed pigmentation. Agronomy Journal 50: 733–734. [Google Scholar]
- Montenegro JD, Golicz AA, Bayer PE, Hurgobin B, Lee H, Chan C‐KK, Visendi P, Lai K, Doležel J, Batley J et al. 2017. The pangenome of hexaploid bread wheat. The Plant Journal 90: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Morgante M, De Paoli E, Radovic S. 2007. Transposable elements and the plant pan‐genomes. Current Opinion in Plant Biology 10: 149–155. [DOI] [PubMed] [Google Scholar]
- Nakamura S. 2018. Grain dormancy genes responsible for preventing pre‐harvest sprouting in barley and wheat. Breed Science 68: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson‐Ehle H. 1914. Zur Kenntnis der mit der keimungsphysiologie des weizens in zusammenhang stehenden inneren faktoren. Zeitschrift Für Planzenzüctung 2: 153–187. [Google Scholar]
- Paterson AH, Sorrells ME. 1990. Inheritance of grain dormancy in white‐kernelled wheat. Crop Science 30: 25–30. [Google Scholar]
- Ransom JK, Bezonsky WA, Sorenson BK. 2006. Hard white wheat: producing North Dakota’s next market opportunity . North Dakota State University Extension Service, Fargo, ND. [WWW document] URL https://www.ag.ndsu.edu/publications/crops/hard‐white‐wheat‐producing‐north‐dakotas‐next‐market‐opportunity [accessed January 2015].
- Rasul G, Humphreys DG, Brûlé‐Babel A. 2009. Mapping QTLs for pre‐harvest sprouting traits in the spring wheat cross ‘RL4452/AC Domain’. Euphytica 168: 363–378. [Google Scholar]
- Rathjen JR, Strounina EV, Mares DJ. 2009. Water movement into dormant and non‐dormant wheat (Triticum aestivum L.) grains. Journal of Experimental Botany 60: 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roistacher CN, Klotz LJ, Eaks IL. 1957. Ammonia gas used in citrus packing plants as fumigant for control of blue‐greenmold on Valencias, navels and lemons. California Agriculture 11: 11–12. [Google Scholar]
- Saxena RK, Edwards D, Varshney RK. 2014. Structural variations in plant genomes. Briefings in Functional Genomics 13: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl SV, Katche E, Ihien E, Chawla HS. 2018. The role of genomic structural variation in the genetic improvement of polyploid crops. Crop Journal 7: 127–140. [Google Scholar]
- Shewry PR, Hey SJ. 2015. The contribution of wheat to human diet and health. Food Energy Secure 4: 178–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. 1995. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. The Plant Journal 8: 659–671. [DOI] [PubMed] [Google Scholar]
- Stetter MG, Vidal‐Villarejo M, Schmid KJ. 2020. Parallel seed color adaptation during multiple domestication attempts of an ancient new world grain. Molecular Biology and Evolution 37: 1407–1419. [DOI] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch SR. 2006. Caught red‐handed: Rc encodes a basic helix‐loop‐helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SM, Xi HX, Ai KJ, Hou HS. 2016. Overexpression of wheat TaNCED gene in Arabidopsis enhances tolerance to drought stress and delays seed germination. Biologia Plantarum 61: 64–72. [Google Scholar]
- Uauy C. 2017. Wheat genomics comes of age. Current Opinion in Plant Biology 36: 142–148. [DOI] [PubMed] [Google Scholar]
- Vetch JM, Stougaard RN, Martin JM, Giroux MJ. 2019. Review: revealing the genetic mechanisms of pre‐harvest sprouting in hexaploid wheat (Triticum aestivum L.). Plant Science 281: 180–185. [DOI] [PubMed] [Google Scholar]
- Volis S. 2016. Seed heteromorphism in Triticum dicoccoides: association between seed positions within a dispersal unit and dormancy. Oecologia 181: 401–412. [DOI] [PubMed] [Google Scholar]
- Walker‐Simmons M. 1988. Enhancement of ABA responsiveness in wheat embryos by high temperature. Plant, Cell & Environment 11: 769–775. [Google Scholar]
- Walkowiak S, Gao LL, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez‐Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D et al. 2020. Multiple wheat genomes reveal global variation in modern breeding. Nature 588: 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, Luo M‐C, Chen ZX, You FM, Wei YM, Zheng YL, Dvorak J. 2013. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D‐genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytologist 198: 925–937. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang XL, Meng JY, Zhang YJ, He ZH, Yang Y. 2016. Characterization of Tamyb10 allelic variants and development of STS marker for pre‐harvest sprouting resistance in Chinese bread wheat. Molecular Breeding 36: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner RL, Kudrna DA, Spaeth SC, Jones SS. 2000. Dormancy in white‐grain mutations of Chinese Spring wheat (Triticum aestivum L.). Seed Science Research 10: 51–60. [Google Scholar]
- Yang J, Liu Y, Pu Z, Zhang L, Yuan Z, Chen G, Wei Y, Zheng Y, Liu D, Wang JR. 2014. Molecular characterization of high pI α‐amylase and its expression QTL analysis in synthetic wheat RILs. Molecular Breeding 34: 1075–1085. [Google Scholar]
- Yang J, Tan C, Lang J, Tang H, Hao M, Tan Z, Yu H, Zhou Y, Liu ZH, Li ML et al. 2019. Identification of qPHS.sicau‐1B and qPHS.sicau‐3D from synthetic wheat for pre‐harvest sprouting resistance wheat improvement. Molecular Breeding 39: 132–143. [Google Scholar]
- Yu M, Chen GY, Zhang LQ, Liu YX, Liu DC, Wang JR, Pu ZE, Zhang L, Lan XJ, Wei YM et al. 2014. QTL mapping for important agronomic traits in synthetic hexaploid wheat derived from Aegilops tauschii ssp. tauschii . Journal of Integrative Agriculture 13: 1835–1844. [Google Scholar]
- Zhang D, He J, Huang L, Zhang C, Zhou Y, Su Y, Li S. 2017. An advanced backcross population through synthetic octaploid wheat as a “bridge”: development and QTL detection for seed dormancy. Fronteir in Plant Science 8: 2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LC, Dong CH, Chen ZX, Gui LX, Chen C, Li DP, Xie ZC, Zhang Q, Zhang XY, Xia C et al. 2021. WheatGmap: a comprehensive platform for wheat gene mapping and genomic studies. Molecular Plant 14: 187–190. [DOI] [PubMed] [Google Scholar]
- Zhang LQ, Liu DC, Yan ZH, Lan XJ, Zheng YL, Zhou YH. 2004. Rapid changes of microsatellite flanking sequence in the allopolyploidization of new synthesized hexaploid wheat. Science in China. Series C, Life Sciences 47: 553–561. [DOI] [PubMed] [Google Scholar]
- Zhong RQ, Ye ZH. 2012. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant and Cell Physiology 53: 368–380. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Chen ZX, Cheng MP, Chen J, Zhu TT, Wang R, Liu Y, Qi P, Chen G, Jiang Q et al. 2018. Uncovering the dispersion history, adaptive evolution and selection of wheat in China. Plant Biotechnology Journal 16: 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tang H, Cheng MP, Dankwa KO, Chen ZX, Li ZY, Gao S, Liu YX, Jiang QT, Lan XJ et al. 2017. Genome‐wide association study for pre‐harvest sprouting resistance in a large germplasm collection of Chinese wheat landraces. Fronteir in Plant Science 8: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Wang SX, Zhao LX, Zhang DX, Hu JB, Cao XL, Yang YJ, Chang C, Ma CX, Zhang HP. 2014. Exploring molecular markers of pre‐harvest sprouting resistance gene using wheat intact spikes by association analysis. Crop Journal 10: 1725–1732. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 KASP markers were developed by SNPs in the QPHS.sicau‐3D (PHS‐3D) region.
Fig. S2 STS markers were developed in the 2.4 Mb PAG region on 3DL for genotyping wheat lines.
Fig. S3 Expression profiles of genes in the PAV region containing PHS‐3D.
Fig. S4 RNA in situ hybridization of Myb10‐D in transversal sections 5‐15 DPA indicates its expression in the mesocarp of the inner pericarp.
Fig. S5 Subcellular localization of Myb10‐D in the nucleus.
Fig. S6 Characterization of binding sites based on the 2‐kb promoter regions of potential Myb10‐D regulated genes in the MEP pathway.
Methods S1 DNA extraction and genotyping.
Methods S2 Fine mapping of PHS‐3D (QPHS.sicau‐3D).
Methods S3 Optical map construction.
Methods S4 Gene cloning and sequencing.
Methods S5 RNA preparation and RNA sequencing.
Methods S6 NGS data analysis (reads mapping, assembly, and functional annotation).
Methods S7 Quantitative PCR with reverse transcription (RT)‐PCR.
Methods S8 Gene transformation.
Methods S9 Endogenous ABA profiling.
Methods S10 Flavonoids extraction and profiling (multiple reaction monitoring, MRM).
Methods S11 Subcellular localization of Myb10‐D.
Methods S12 RNA in situ hybridization.
Methods S13 Protein expression in Escherichia coli and purification.
Methods S14 Electrophoretic mobility shift assay.
Methods S15 Transient expression assay.
Methods S16 One‐hybrid assays in yeast.
Methods S17 Statistical analyses.
Methods S18 Code used in this study.
Table S1 Primers used for KASP, STS, PCR clone, qPCR, transgenetic, and transactivation assay.
Table S2 Genotyping and phenotyping of 262 common wheat, 16 Aegilops tauschii, and three tetraploid wheat lines.
Table S3 Summary of genomic reference sequences data and NGS data obtained in this study (NCBI, PRJNA605937).
Table S4 Genes in the 2.4 Mb PAV region from six accessions CS, AK58, AL8/78, Robigus, Cadenza, and SHW‐L1.
Table S5 Transposons in the 2.4 Mb PAV region from CS and AK58.
Table S6 The expression of 20 genes in 2.4 Mb PAV region from CM32 and SHW‐L1 in 10‐30 DPA seeds, and the expression of three selected genes from CS and BCScv1 in 29 tissues or stages.
Table S7 Germination percentages of WT and OE lines.
Table S8 Different expression genes that had similar expression patterns with Myb10‐D form WT and OE lines in 5‐30 DPA seeds.
Table S9 Accumulation of flavonoids and ABA in WT and OE lines.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The sequence data that support the findings of this study are openly available in NCBI Sequence Read Archive at https://www.ncbi.nlm.nih.gov/, under accession number PRJNA605937. Other main data that supports the findings of this study are available in the Supporting Information of this article.


