Figure 2.
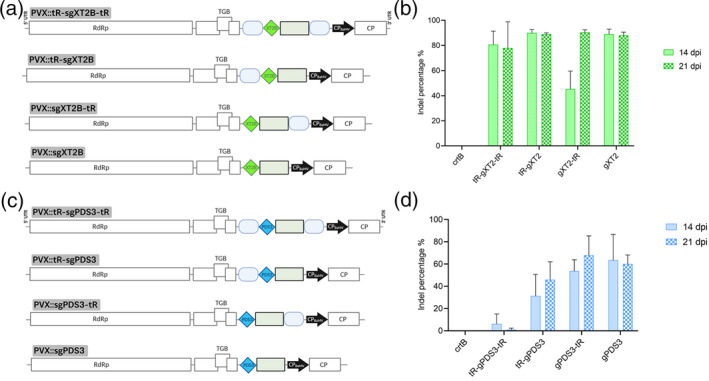
Single gene editing, with or without flanking tRNAs, using the PVX‐based system in Nicotiana benthamiana.
(a) Schematic representation of recombinant clones PVX::tR‐sgXT2B‐tR, PVX::tR‐sgXT2B, PVX::sgXT2B‐tR and PVX::sgXT2B. NbXT2B protospacer is represented by a green diamond. Other details as in the legend to Figure 1.
(b) Inference of CRISPR edits (ICE) analysis of the first systemically infected upper leaf of N. benthamiana plants inoculated with PVX::sgXT2B derivatives at 14 days post‐inoculation (dpi) (n = 3) and 21 dpi (n = 3). PVX::crtB was used as a negative editing control.
(c) Schematic representation of recombinant clones PVX::tR‐sgPDS3‐tR, PVX::tR‐sgPDS3, PVX::sgPDS3‐tR and PVX::sgPDS3. NbPDS3 protospacer is represented by a blue diamond. Other details as in the legend to Figure 1.
(d) ICE analysis of the first systemically infected upper leaf of N. benthamiana plants inoculated with PVX::sgPDS3 derivatives at 14 dpi (n = 3) and 21 dpi (n = 3). (b,d) Columns and error bars represent average indels (%) and standard deviation, respectively.
