Figure 2.
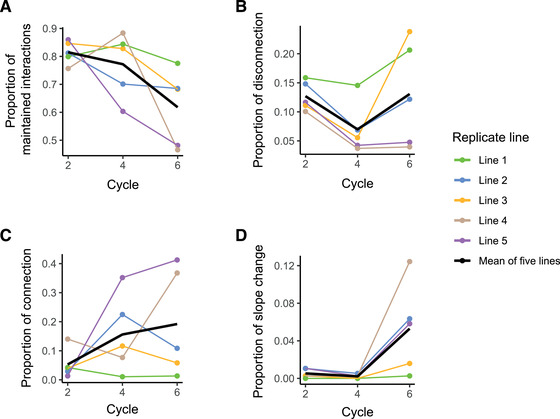
. Rapid rewiring of trait correlation networks during evolution of Pseudomonas protegens CHA0 in the rhizosphere of gnotobiotic Arabidopsis thaliana Col‐0. The systematic classification scheme for traits correlation patterns has been presented in Methods and Figure 1. (A) We defined maintained bacterial trait interactions as the fraction of the pairwise trait combinations that either remained with no correlation or kept significant association with unchanged correlation coefficient in the evolved population as compared to the ancestral population (ANOVA, nonsignificant Slope × Time interaction). (B–D) Proportion of pairs of traits in which we observed innovation, defined as significant changes of pairwise variable interactions, including disconnection (B, loss of significant correlation), connection (C, emergence of a significant correlation), and slope change (D, significant change in slope). Cycle numbers on the x‐axis represent the number of plant growth cycles, with bacteria being transferred to a new plant at the end of each growth cycle. Different colors indicate the five replicate lines, and the black line represents the average of all lines. In total, (28 × 27)/2 = 378 pairs of traits were tested for their interaction conservativism. The distinction between association and no association is made based on statistical significance. P‐values were adjusted with Benjamini‐Hochberg (BH) procedure.
