Fig. 6.
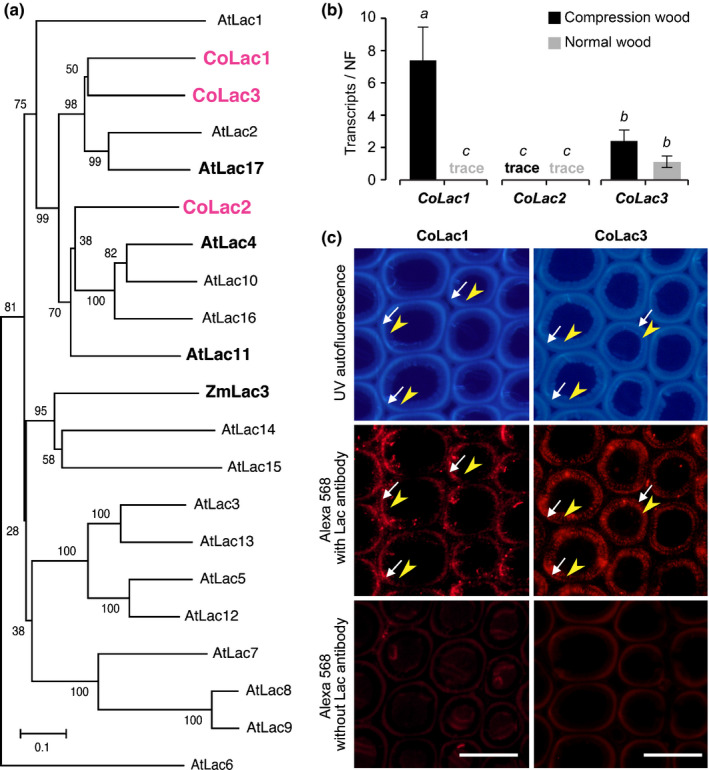
Laccases responsible for lignification in Chamaecyparis obtusa compression wood. (a) Phylogenetic analysis of laccase proteins. An unrooted phylogenetic tree was built using neighbour‐joining method. Bootstrapping with 1000 replicates was performed. Bar, 0.1 substitutions per site. (b) Transcript abundance of CoLac1, CoLac2 and CoLac3 in differentiating compression and normal wood tracheids of C. obtusa seedlings as determined using quantitative real‐time PCR. The normalisation factor (NF) for calibration of transcript abundance was determined as described in Methods S1. Values are the mean ± SD for three independently analysed plants. Different letters indicate significant differences (ANOVA with post‐hoc Tukey–Kramer test, P < 0.05). (c) Immunolocalisation of CoLac1 and CoLac3 in compression wood cell walls of C. obtusa seedlings. Sections were visualised using autofluorescence from deposited lignin and Alexa 568 secondary antibody fluorescence from CoLac1 and CoLac3 epitopes. Control sections visualised without anti‐CoLac1 or anti‐CoLac2 treatment are also shown. White arrows and yellow arrowheads indicate the S2L and iS2 layers, respectively. Bars, 20 μm.
