Fig. 7.
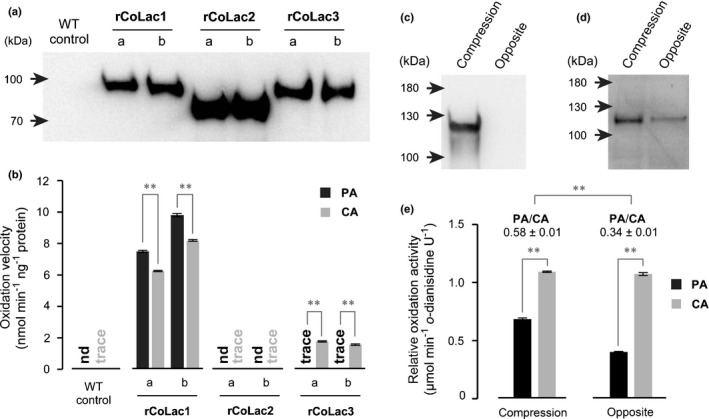
Enzyme assays for recombinant and native Chamaecyparis obtusa laccases. (a, b) Recombinant C. obtusa laccase proteins, rCoLac1, rCoLac2 and rCoLac3, were expressed in tobacco BY‐2 cells. Cell‐wall‐bound protein fractions prepared from two independent transformed lines for each construct (a, b) and control wild‐type cell lines (WT) were used for the laccase activity assay using PA and CA as substrates. Western blots for protein detection using an anti‐His6 antibody (a) and monolignol‐oxidation velocities of rCoLac1, rCoLac2 and rCoLac3 protein preparations (b) are shown. Data in (b) are expressed as the mean ± SD for three independent reaction runs. Asterisks indicate significant difference between PA and CA oxidation velocities (Student’s t‐test; **, P < 0.01). nd, not detected. (c–e) Monolignol‐oxidation activities of cell‐wall‐bound protein fractions prepared from differentiating compression wood tissues of a bent mature C. obtusa tree. Protein fractions from normal‐wood‐forming opposite wood tissues of the same tree were used as a control. Western blots using anti‐CoLac1 (c) and anti‐CoLac3 (d) antibodies, and relative monolignol‐oxidation activities of the cell‐wall‐bound protein fractions (e) are shown. Data in (e) are expressed as oxidation velocities relative to o‐dianisidine used as a reference substrate, and values are the mean ± SD for three independent reaction runs. Asterisks indicate significant difference between PA and CA oxidation velocities and PA/CA oxidation ratios between the compression wood and opposite wood control proteins (Student’s t‐test; **, P < 0.01).
