FIGURE 2.
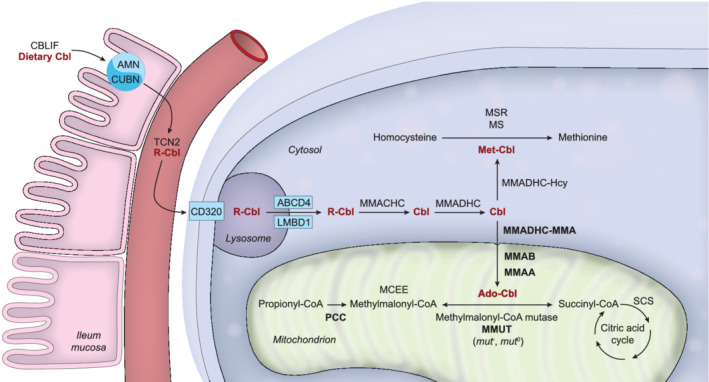
Scheme depicting the extra‐ and intracellular transport and processing of the cobalamin (Cbl) molecule and the involved proteins. Methylmalonyl‐CoA mutase (MMUT), methylmalonic aciduria type A protein (MMAA), methylmalonic aciduria type B protein (MMAB), methylmalonic aciduria and homocystinuria type D protein variant 2 (MMADHC‐MMA), and propionyl‐CoA carboxylase (PCC) defects and their related diseases are discussed in these guidelines, hence these proteins are depicted in bold print. The other proteins are depicted using official protein nomenclature: amnionless (AMN) and cubulin (CUBN) form the cubam receptor to absorb cobalamin bound to cobalamin binding intrinsic factor (CBLIF); haptocorrin (TCN1; not shown), transcobalamin (TCN2) and transcobalamin receptor (CD320) facilitate cobalamin transport and uptake into the cell; lysosomal cobalamin transporter (ABCD4) and lysosomal cobalamin transport escort protein (LMBD1) export cobalamin from the lysosome; methylmalonic aciduria and homocystinuria type C protein (MMACHC) cleaves the R group of cobalamin (upper‐axial ligand); methylmalonic aciduria and homocystinuria type D protein (MMADHC) targets cobalamin towards further processing in the cytosol or the mitochondrion; methionine synthase (MS), kept in its active form by methionine synthase reductase (MSR), uses cobalamin in its methylated form; methylmalonyl‐CoA epimerase (MCEE) converts (R)‐methylmalonyl‐CoA to (S)‐methylmalonyl‐CoA; succinyl‐CoA synthetase (SCS) also called succinate‐CoA ligase forms succinate from succinyl‐CoA in the citric acid cycle
