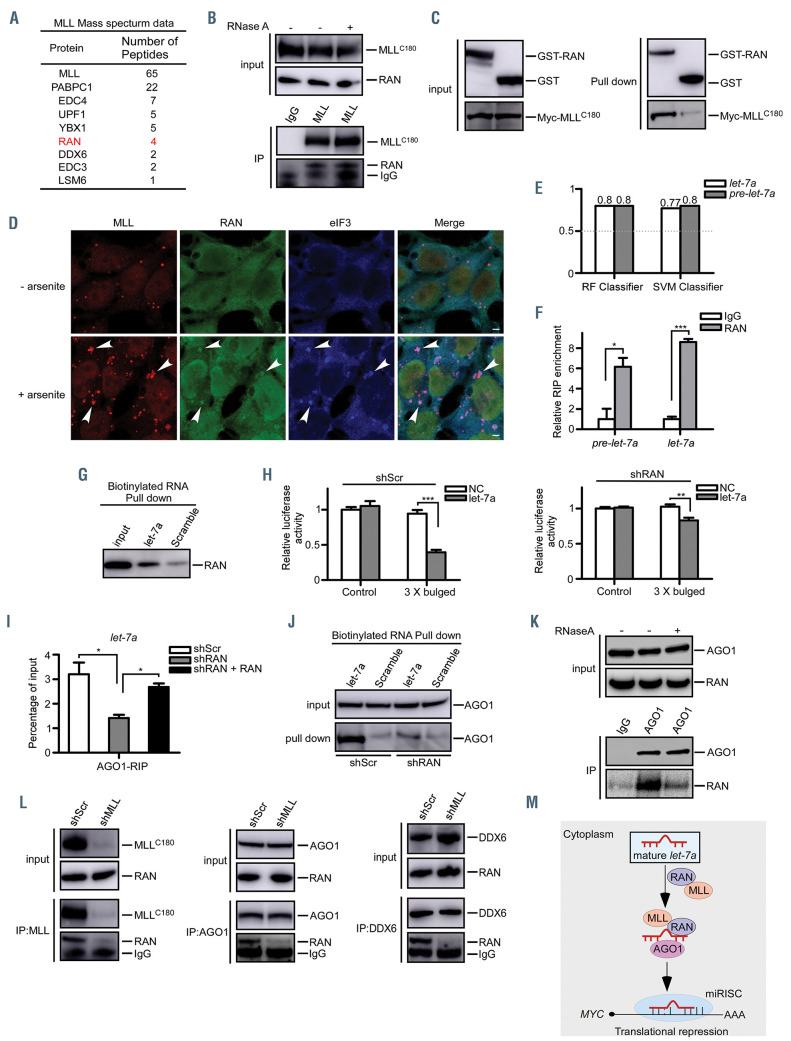
Figure 2.
MLL contributes to the loading of let-7a onto AGO1 through interacting with RAN. (A) List of MLL-associated proteins identified by mass spectrometric analysis. 293T cells transfected with MLL were harvested and subjected to the nuclear-cytoplasmic fractionation. The cytoplasmic fractions were prepared for the immunoprecipitation assays followed by mass spectrometric analysis. (B) 293T cell lysates were treated with RNase A followed by anti-MLL immunoprecipitation. Western blots were performed using the indicated antibodies. (C) Direct interaction between MLLC180 and GST-RAN was examined. Left panels: western blots showing the inputs of purified GST-RAN and Myc-MLLC180. Right panels: the pull-down immunoblots were shown with GST-RAN as the bait and the pulled MLLC180 detected by an anti-Myc antibody. (D) 293T cells were untreated (upper panels) or treated with arsenite (0.5 mM, 45 min) (lower panels), then fixed and stained with the indicated antibodies. Note that eIF3 is specific for stress granules. Arrowheads show the localization of MLL with RAN and eIF3. Scale bar, 5 mm. (E) The RPISeq tool was used to predict the interactions between RAN and let-7a or pre-let-7a. The random forest (RF) classifier and support vector machine (SVM) classifier represent the confidence of the prediction. In performance evaluation experiments, predictions with probabilities >0.5 were considered “positive”. (F) 293T cellular lysates were prepared and anti-RAN RIP experiments were performed. Pulled down RNA were isolated, pre-let-7a and mature let-7a were analyzed by qRT-PCR using specific primers. (G) 293T cellular lysates were subjected to biotinylated- let-7a RNA pull-down assays. Then let-7a-immunoprecipitated RAN proteins were subjected to western blot analysis. Scrambled miRNA were used as negative controls. (H) 293T-shScr and shRAN cells transfected with Agomir-negative control (NC) or Agomir- let- 7amimic (let-7a) were subjected to dual luciferase reporter assays. The ratio of luciferase activity was measured and normalized to the value of the cells transfected with the control reporter and NC. (I) Extracts of 293T-shScr and shRAN cells, with the latter being rescued by shRNA-resistant RAN, were subjected to anti-AGO1 RIP assays. Pulled-down RNA were analyzed by qRT-PCR using specific primers for let-7a. (J) 293T-shScr and 293T-shRAN cellular lysates were subjected to biotinylated- let-7a RNA pull-down assays. Then let-7a-immunoprecipitated AGO1 proteins were subjected to western blot analysis. Scrambled miRNA were used as negative controls. (K) 293T cell lysates were treated with RNase A followed by anti-AGO1 immunoprecipitation. Western blots were performed using the indicated antibodies. (L) Extracts of 293T-shScr and 293T-shMLL cells were collected and co-immunoprecipitation assays were performed and analyzed using the indicated antibodies. (M) The proposed mechanism through which MLL and RAN are involved in the loading of let-7a onto AGO1. MLL is required for the loading of let-7a onto AGO1 via a direct interaction with RAN. Thus, RAN serves as a molecular adaptor for the assembly of MLL-associated miRISC. NS, no significant difference. *P<0.05, **P<0.01, ***P<0.001. Data represent the mean and standared error of mean of three independent experiments.