Fig. 5.
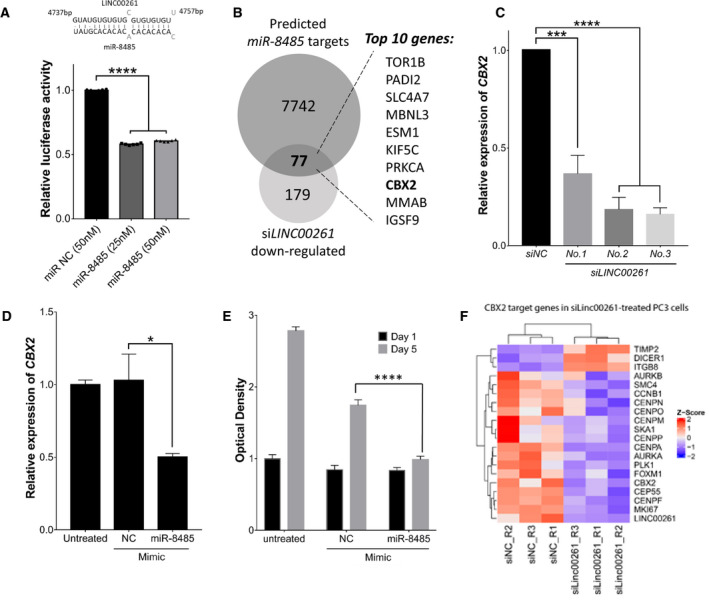
Cytoplasmic LINC00261 buffers miR‐8485 increasing CBX2 activity to drive proliferation in NEPC cells. (A, top) Computational analyses predict miR‐8485 binding within 3′UTR of LINC00261. The partially complementary seed sequence is visualized below. (A, bottom) Luciferase activity from the LINC00261‐binding reporter assay with transfection of miR‐8485 or a nontargeting control miRNA mimic in HEK293 cells. For each treatment group, reporter activity was normalized to background signals from the unmodified pmirGlo reporter alone. (B) Overlap between predictive targets of miR‐8485 (n = 7742) and differentially expressed genes (DEGs; n = 179) in LINC00261‐silenced PC‐3 cells (RNA‐seq). 77 gene candidates were common in the two lists with top 10 genes shown. (C) CBX2 expression (qPCR) in PC‐3 cells treated with a nontargeting control (siControl) or siRNA targeting LINC00261 (***P = 0.0002; ****P < 0.0001) or (D) treated with miR‐8485 or nontargeting miRNA mimic (*P = 0.0434). (E) Viability (MTT assay) of PC‐3 cells treated with a negative control mimic (NC mimic) or miR‐8485 mimic (****P < 0.0001). (F) Heat map showing mRNA expression (RNA‐seq) changes in CBX2 target genes in PC‐3 cells treated with siLINC00261 or siNC. (A, B) Analyzed by two‐tailed t‐test n = 6. (D) Analyzed by one‐way ANOVA with Dunnett's post hoc test n = 2. (E) Analyzed by two‐tailed t‐test n = 3. (F) Analyzed by two‐way ANOVA with Tukey's post hoc test n = 6. All data are shown as mean ± SEM.
