ABSTRACT
The idea of enhanced methanol production from cell wall by pectin methyl esterase enzymes (PME) combined with expression of cry genes from Bacillus thuringiensis as a strategy to improve insect pest control in cotton is presented. We constructed a cassette containing two cry genes (cry1Fa and Cry32Aa) and two pme genes, one from Arabidopsis thaliana (AtPME), and other from Aspergillus. niger (AnPME) in pCAMBIA1301 plant expression vector using CAMV-35S promoter. This construction was transformed in Eagle-2 cotton variety by using shoot apex-cut Agrobacterium-mediated transformation. Expression of cry genes and pme genes was confirmed by qPCR. Methanol production was measured in control and in the cry and pme transformed plants showing methanol production only in transformed plants, in contrast to the non-transgenic cotton plants. Finally, insect bioassays performed with transgenic plants expressing cry and pme genes showed 100% mortality for Helicoverpa armigera (cotton bollworm) larvae, 70% mortality for Pectinophora gossypiella (pink bollworm) larvae and 95% mortality of Earias fabia, (spotted bollworm) larvae, that was higher than the transgenic plants expressing only cry genes that showed 84%, 49% and 79% mortality, respectively. These results demonstrate that Bt. cry-genes coupled with pme genes are an effective strategy to improve the control of different insect pests.
KEYWORDS: Cotton, transformation, eagle-2, agrobacterium, pectin methyl esterase enzyme, insecticidal cry proteins
1. Introduction
Gossypium hirsutum L. is an important economical crop and one of the largest sources of natural fiber worldwide.1 Cotton crop is subjected to various biotic and abiotic stresses throughout its life cycle.2 However, the biotic stress caused by pathogens and pests has an important negative impact not only on yield and quality, but also in control measures that increases production cost globally.1 Cotton fields are prone for lepidopteran infestation such as pink bollworm (Pectinophora gossypiella), army bollworm (Spodoptera litura), American bollworm (Heliothis armigera) and spotted bollworm (Earias fabia). The use of chemical insecticides is not an adequate solution since they seriously damage the environment and human health.3 Bacillus thuringiensis (Bt) is a soil-born bacterium that produces different insecticidal proteins (named δ-endotoxins, such as Cry toxins), which have been successfully used against insect pest attacks and several Cry insecticidal proteins have been also transformed into cotton crops since 1996.4 The effectiveness of Bt δ-endotoxins started to decline due to the development of resistance by the insect pests.5 Despite the proven substantial protective effects of transgenic Bt-cotton plants against insect attack, improvement is still needed in this technology,6 for instance by combining with some enzymes involved in defense strategies against insect pests.7 The overproduction of enzymes, involved in insect defense, can be a good alternative to reduce the pest attack and development of insect resistance to Cry toxins.7
Plant cell walls are heterogeneous structures containing cellulose, hemicellulose, pectin, phenolic compounds and cell wall proteins. Pectins are integral components of plant primary cell wall acting as barriers to insect pests. Pectin methyl esterase enzyme (PME) catalyzes desterification of pectin into pectate and methanol in the plant cell wall to regulate an inhibitory response against insect pests.3,8 Multiple mechanisms are involved in regulation of PME activity and methanol production in plants such as cell wall pH modifications, expression of inhibitory proteins and differential isoform expression in different tissues at different stages.9
In this work, PME from Arabidopsis thaliana (AtPME, accession no. NP 566842) and Aspergillus niger (AnPME, accession no. XM_001390469) were used for their overexpression coupled with insecticidal cry genes in transgenic cotton and toxicity was evaluated against different Lepidopteran insect pests. We selected to work with the PME isolated from the fungus A. niger shows high methanol production in tobacco cell suspension culture.10 It is also reported that activity of PME is elicited by the attack of herbivores in different plant species,7,11,12 inducing the production of toxic methanol.13 Methanol is toxic to insect pests,14 damaged and wounded leaves by the insect attack as a primary source of methanol production by the preexisting PME in the cell wall.15
2. Materials and Methods
2.1. Plant Materials
Gossypium hirsutum L. Eagle-2 variety was selected for the expression of two cry genes (cry1Fa and cry32Aa) and two methanol producing genes (AtPME and AnPME). Eagle-2 seeds were obtained from Four Brothers Seeds Multan-Pakistan and planted at research farm of Four Brothers Lahore-Pakistan.
2.2. Sequence Selection and Plasmid Construction
Gene sequences of selected pme genes (AtPME, accession no. NP 566842; AnPME, accession no. XM_001390469) were taken from NCBI and after codon optimization, were synthesized by BioBasic, Canada. The synthetic double gene AnPME, AtPME cassette (total 7893 bp) was cloned into the EcoRI and HindIII restriction sites of puc57 vector. While the cry1Fa, cry32Aa gene cassette (total 7954 bp) was cloned into the EcoRI and BamHI restriction sites of pUC57 vector. All genes were under regulation of CaMV35S promoter and Nos terminator was added at the end of these genes (Fig. 1).
Figure 1.
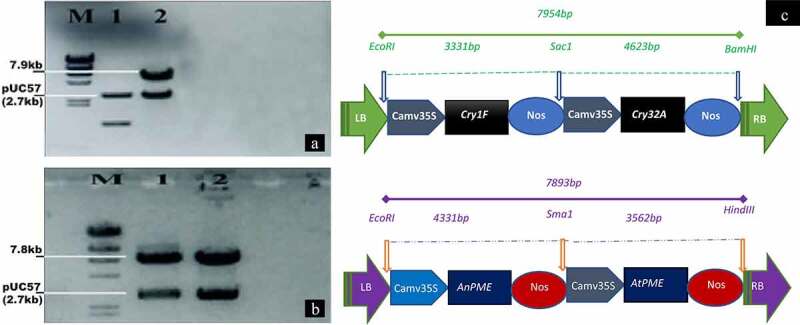
Restriction analysis of puc57 vectors containing the cry1Fa and cry32Aa gene cassette (total 7954 bp size) or the AtPME and AnPME gene cassette (total 7893 bp size). Figure 1a, BamHI and EcoRI restriction analysis of the cry1Fa and cry32Aa gene cassette. M: 10kb ladder. Lane 2: cry1Fa and cry32Aa gene cassette positive sample. Lane 1: negative control of pUC57 without insert samples. Figure 1b, EcoRI and HindIII restriction analysis of the AtPME and AnPME gene cassette. M: 10 Kb ladder. Lane 1, 2: AtPME and AnPME gene cassette positive samples. Figure 1c, schematic representation of both gene cassettes containing cry1Fa and cry32Aa genes or AtPME and AnPME genes, respectively
2.3. Cloning of Genes into Plant Expression Vector pCAMBIA1301
The two pUC57 vectors with the AtPME, AnPME cassette, or the cry1Fa, cry32Aa cassette were transformed into Escherichia coli top10 competent cells through heat shock method and selected on LB media supplemented with tetracycline (50 µg/ml) and ampicillin (50 µg/ml). Plasmids were isolated by using Gene Jet plasmid DNA isolation kit (Thermo Scientific, Vilnius, Lithuania, Cat#K0503) as indicated in the manufacturer protocol. The presence of these genes was confirmed through restriction digestion using EcoRI and HindIII or using EcoRI and BamHI enzymes accordingly to each plasmid. The 7.8 DNA band of AtPME, AnPME cassette and 7.9 Kb band of cry1Fa, and cry32Aa cassette were visualized in 0.8% of agarose gel electrophoresis and excised fragments were purified by using Gene JET Gel Extraction Kit (Thermo Scientific, Vilnius, Lithuania, Cat#K0503). The purified DNA bands were then ligated to the plant expression vector pCAMBIA1301 pre-digested with the corresponding restriction enzymes and transformed into E. coli top10 competent cells. A 1Kb DNA size marker (250bp-10Kb) was received from GeneRulerTM (Thermo Scientific, Cat#SM0311) and used in this study.
2.4. Transformation of pCAMBIA1301 vectors with pme and cry gene cassettes into Agrobacterium tumefaciens.
The ligation of the two excised double gene cassettes in pCAMBIA1301 vector was confirmed by restriction digestion using EcoRI and BamHI enzymes or EcoRI and HindIIII accordingly to the pCAMBIA1301-cryCassette or to the pCAMBIA1301-pmeCassette, and further confirmed by PCR using specific primers (see Table 1 and 4)
Table 1.
Sequences of the primers used
| Primers | Sequence |
| GADPH-F | TGATGCCAAG GCTGGAATTGCTT |
| GADPH-R | GTGTCGGATCAAGTCGATAACACGG |
| AnPME-F | GGTGCTATCGTTGTTGCTAAGTC |
| AnPME-R | GCAGTAATTGAAGCAGATGAAGG |
| AtPME -F | TCTGTTCCTTTGGGTAACACTTG |
| AtPME -R | GTGATCACGCACCTAAGAAAGAC |
| Cry1Fa-F | ATGCGCTGTTTACTTCTAGCAAC |
| Cry1Fa-R | GCACATTTACTGTTTCGTGTTTG |
| Cry32a-F | CTTTTGCTACGGCTGGACTC |
| Cry32a-R | TCACTGCCTTTCTGGCTTTT |
| VirG-F | GAATACCTTACGATCCACGCC |
| VirG-R | GCGAAACCTGCACGTCCGCG |
Table 2.
Numerical data for transformation experiments in the field
| Exp. No. | No. of embryos isolated | Agrobacterium- treated embryos |
Embryos on MS plates | Died | Selection tubes | Plantlets died | Plants transferred to pots | plants died in pots | Plants shifted to green house |
| 1 | 403 | 403 | 403 | 396 | 7 | 1 | 6 | 1 | 5 |
| 2 | 345 | 345 | 345 | 341 | 4 | 2 | 2 | 1 | 1 |
| 3 | 314 | 311 | 311 | 307 | 4 | 2 | 2 | 1 | 1 |
| 4 | 310 | 305 | 305 | 298 | 7 | 1 | 6 | 3 | 3 |
| 5 | 350 | 345 | 345 | 340 | 5 | 2 | 3 | 0 | 3 |
| 6 | 409 | 410 | 410 | 404 | 6 | 2 | 4 | 2 | 2 |
| 7 | 320 | 319 | 319 | 314 | 5 | 2 | 3 | 1 | 2 |
| 8 | 410 | 410 | 410 | 404 | 6 | 3 | 3 | 1 | 2 |
| 9 | 311 | 311 | 311 | 309 | 2 | 1 | 1 | 0 | 1 |
| 10 | 336 | 335 | 335 | 330 | 5 | 2 | 3 | 1 | 2 |
| 11 | 400 | 410 | 410 | 406 | 4 | 1 | 3 | 2 | 1 |
| 12 | 315 | 312 | 312 | 310 | 2 | 1 | 1 | 1 | 0 |
| Total | 4223 | 4216 | 4216 | 4159 | 57 | 20 | 37 | 14 | 23 |
Table 3.
Germination Index
| No. of petri plates | Total seeds | No. of germinated seeds | No. of ungerminated seeds | Germination index |
| 1 | 40 | 27 | 21 | 67.50% |
| 2 | 40 | 31 |
Table 4.
Transformation efficiency
| Agrobacterium treated embryos |
Control plants | Plants shifted to green house | transformation efficiency |
| Control plants Experimental | Control plants Experimental | ||
| 4216 | 50 | 23 57 | 46% 1.35% |
The optimized PCR conditions used for Cry genes; Initial denaturation at 94°C for 3 minutes, denaturation at 94°C for 45 seconds, annealing at 59°C for 50 seconds, extension at 72°C for 1:30 minutes, final extension at 72°C for 10 minutes whereas the PCR conditions for pme genes; Initial denaturation at 94°C for 3 minutes, denaturation at 94°C for 45 seconds, annealing at 57°C for 45 seconds, extension at 72°C for 1:30 minutes, final extension at 72°C for 10 minutes. The confirmed plasmids were transformed into A. tumefaciens strain LBA4404 competent cells by electroporation.16 The transformant A. tumefaciens cells were grown on YEP media (Peptone 10 g/L, Yeast extract 10 g/L, Sodium chloride 5 g/L, pH 7.5) supplemented with Kanamycin (50 mg/ml) and Rifampicin (50 mg/ml). The appeared colonies of A. tumefaciens were then further evaluated through colony PCR as been previously reported17 using the specific primers in Table 1.
2.5. Agrobacterium Tumefaciens Mediated Transformation of Cotton Plants
The seeds of Gossypium hirsutum Eagle-2 plant were surface sterilized and placed in the dark at 30°C for 48 h. The germinated seedlings were used for co-transformation using shoot apex cut method. The embryos were isolated, and injury was introduced to the shoot apex before incubation with the Agrobacterium strain LBA4404 harboring the pCAMBIA1301-cry-pme double cassette. The cultures were incubated for 1 hr at 28°C by placing explants on MS solid medium supplemented with (4.4 g/L, Sucrose 30 g/L, Phytagel 2.4 g/L). The embryos were allowed to grow on MS medium plates for 48–72 h supplemented with cefotaxime (500 mg/ml) followed by screening in MS tubes supplemented with hygromycin (25 mg/ml) for one and a half month. After screening, the cotton plants from the tubes were transplanted into pots containing equal proportion of clay, peat moss and sand (1:1:1). Subsequently, the putative transgenic cotton plants were transplanted to the greenhouses of Four Brothers Genetics Inc. for acclimatization and hardening followed by molecular analysis.
2.6. Detection of the Two Double Gene Cassettes in Cotton Plants through PCR
Leaves of the putative transgenic cotton plants were harvested, and DNA was isolated for PCR screening of AnPME, AtPME, cry1Fa, and cry32Aa genes using PCR master mix kit (Thermo Scientific, cat#K1081) using specific primers. In addition, virG gene amplification was also done, by using a specific set of primers from the vir region, to nullify the Agrobacterium contamination. The PCR annealing temperature was set at 60°C.
2.7. RNA Extraction and cDNA Preparation
RNA from putative cotton plants was isolated using Agilent kit (Agilent Technologies, Santa Clara, USA, Cat #5185-6000). The RNA was quantified in ng/µl using NanoDrop ND-1000 spectrophotometer at 260 and 280 nm. The DNase-treated total RNA was used to prepare cDNA using the first strand cDNA synthesis kit (Thermo Scientific, Cat #K1632) and cDNA was stored at −20°C.
2.8. Expression Analysis of Cotton Transgene
Expression analysis of transgenes was performed by qPCR using specific primers in triplicates with a product size of <200 bp following the protocol of Maxima SYBR Green/ROX (Thermo Scientific, Cat#K0221). The reaction mixture was prepared in a total of 20 µl with the following components of 1 µl of 10 pmol of forward and reverse primers, 5 µl of Maxima® SYBR Green/ROX qPCR Master Mix (2x) and 1 µl (50 ng/µl) of cDNA. Sequences of the primers used for the amplifications of both the genes are given in Table 1. Relative expression was determined according to the 2 (-ΔΔCt) method using GAPDH primers were used as internal control reference gene for normalization in these qPCR experiments. All assays were done in triplicate.
2.9. Methanol Quantification in Transgenic Cotton Plants
Transgenic cotton leaves (1 g) were used for determining methanol concentration. Phosphate buffer was prepared in deuterium oxide composed of 0.03% (w/v) sodium salt of trimethylsilyl propionic acid (TSP) (Sigma-Aldrich). After sonication of the samples, they were centrifuged at 13,000 × g for 10 min and supernatant was collected in a tube for methanol content determination, by using mass spectrometry (MS) with the procedure described by.10 Different concentrations of methanol from 0% to 20% were used as standards.
2.10. Insect Bioassays
The efficacy of methanol overproduction was tested on insect bioassays against Helicoverpa armigera, Pectinophora, gossypiella and Earias fabia larvae by comparing toxicity of non-transgenic and transgenic cotton plants. The upper fully expanded positive leaves of the plant that have H. armigera and E. fabia larvae were removed and placed on moist filter paper in laboratory conditions. A 5–7 larvae of 3rd instar were used per leaf in triplicate. The efficacy of transgenic plants against pink bollworm (P. gossypiella) was evaluated by releasing the larvae on young growing bolls and flowers in the field. The mortality rate was observed continuously for 7 days. The mortality rate was calculated by the following formulae;
% Mortality = No. of dead Larvae ∕ Total No. of Larvae × 100
3. Results
3.1. Detection of Pme or Cry Genes Cassettes in pUC57 Vector through Restriction Digestion
The two plasmids pUC57 that we received from BioBasic, were digested with BamHI and EcoRI or with EcoRI and HindIII and resolved on 0.8% agarose gel as shown in Fig. 1a. The expected sizes of 7954 bp and 7893 bp, respectively, for the cassettes containing cry genes or pme gene cassettes were observed (Fig. 1b). A schematic representation of both gene cassettes containing cry1Fa and cry32Aa genes or AtPME and AnPME genes, respectively, is also shown in Fig. 1c.
3.2. Detection of Transgene in pCAMBIA through Restriction Digestion Analysis and PCR
The purified DNA bands containing these two gene cassettes were ligated to the plant expression vector pCAMBIA1301 pre-digested with the corresponding restriction enzymes. The resulting plasmids were confirmed by restriction digestion analysis. Digestion with EcoRI and HindIII was done for the plasmid containing cry gene cassette and digestion with EcoRI and BamHI was done for the plasmid containing pme gene cassette. The observed band sizes confirmed their successful ligation in pCAMBIA1301. Both cry genes were then amplified resulting in a PCR product of 459 bp for cry1F and 462 bp for cry32A. Similarly, AnPME and AtPME genes were amplified through colony PCR resulting in a PCR product of 557 bp and 554 bp, respectively (Fig. 2).
Figure 2.
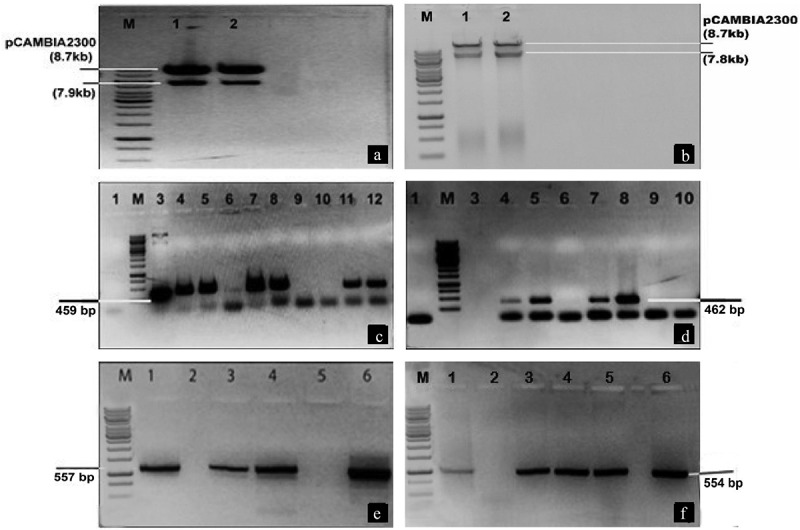
Restriction digestion and PCR analysis of both cry genes and pme genes cassettes
Figure 2a, EcoRI and BamHI restriction of pCAMBIA1301 vector containing cry gene cassette; Lanes 1, 2: show positive samples; Fig. 2b, EcoRI and HindIII restriction of pCAMBIA1301 vector containing pme gene cassette; Lanes 1, 2: show positive samples; Fig. 2c, confirmation of cry1Fa gene by PCR, M: 10 Kb ladder; Lane 1: shows negative control; Lane 3: shows positive control; Lanes 4, 5, 7, 8, 11 and 12: show positive samples; Lanes 6, 9, and 10: show negative samples. Figure 2d, confirmation of cry32Aa gene by PCR, M: 10Kb ladder; Lane 1, 3: shows negative control; Lanes 4, 5, 7: and 8 show positive samples; Lanes 6, 9, and 10: show negative samples. Figure 2e, confirmation of AnPME gene by PCR, M: 1 Kb ladder; Lane 1: shows positive control; Lanes 3, 4, and 6: show positive samples; Lanes 2, and 5: show negative samples. Figure 2f, confirmation of AtPME gene by PCR, M: 10 Kb ladder; Lane 1: shows positive control; Lanes 3, 4, 5, and 6: show positive samples; Lane 2: shows a negative sample.
3.3. Detection of pCAMBIA1301-PMECassette and pCAMBIA1301-cryCassette constructions transformed into A. tumefaciens
A number of random colonies of electroporated A. tumefaciens strain LBA4404 were selected for PCR colony assays, using specific primers designed from cry1Fa, cry32Aa, AnPME and AtPME genes. The PCR products were resolved on 1.5% agarose gel and our results show that all the colonies amplified the expected PCR product of 459 bp and 462 bp for the cry1F and cry32A, respectively, and 557 bp and 554 bp for the Anpme and Atpme genes, respectively, with the exception of the negative control (Fig. 3).
Figure 3.
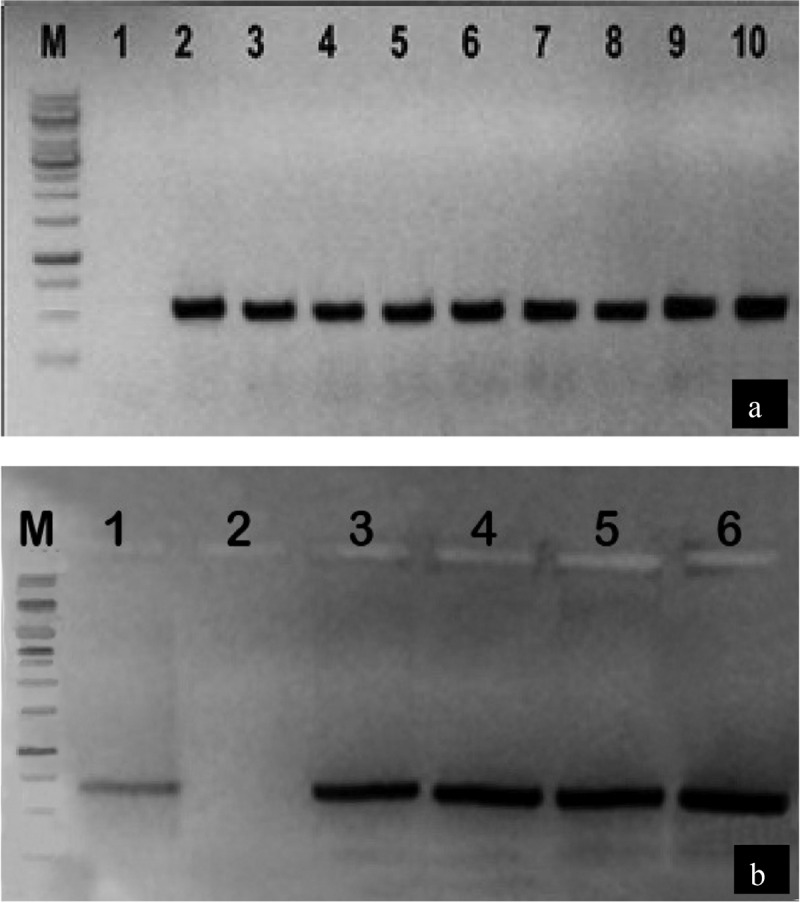
PCR amplification of both cry genes and pme genes from transformed A. tumefaciens with the corresponding pCAMBIA1301-cryCassette and pCAMBIA1301-pmeCassette constructions
Figure 3a, Amplification of both cry genes; M: 10 kb ladder; Lane 1: shows negative control; Lane 2: shows positive control; Lanes 3–6: show positive amplification of cry1Fa, and Lane 7–10: show positive amplification of cry32Aa genes in individual A. tumefaciens colonies. Figure 3b, shows an amplification of both pme genes, M: 10 Kb ladder; Lane 1: shows positive control; Lane 2: shows negative control; Lanes 3, 4: show positive amplification of AnPME, and Lane 5, 6: show positive amplification of AtPME genes in individual A. tumefaciens colonies.
3.4. Transformation of the Two Double Gene Cassettes into Cotton Plants
The seeds of Gossypium hirsutum Eagle-2 were sterilized and placed in the dark at 30°C for 48 h. The germinated seedlings were used for transformation using shoot apex cut method. These plants were inoculated with both A. tumefaciens strains containing cry gene cassette or pme gene cassettes. Germination index of Eagle-2 was calculated to be 67.50% whereas transformation efficiency was recorded to be 1.35%.
3.5. Detection of Transgenic Cotton Plants
Fresh leave samples of the transgenic cotton plants were selected for DNA isolation and analysis of successfully transformed cry genes and pme genes using specific primers that produce PCR products of 459 bp and 462 bp for cry (cry1F & cry32A) genes whereas 557 bp and 454 bp for pme (Anpme & Atpme) genes, respectively (Fig. 4).
Figure 4.
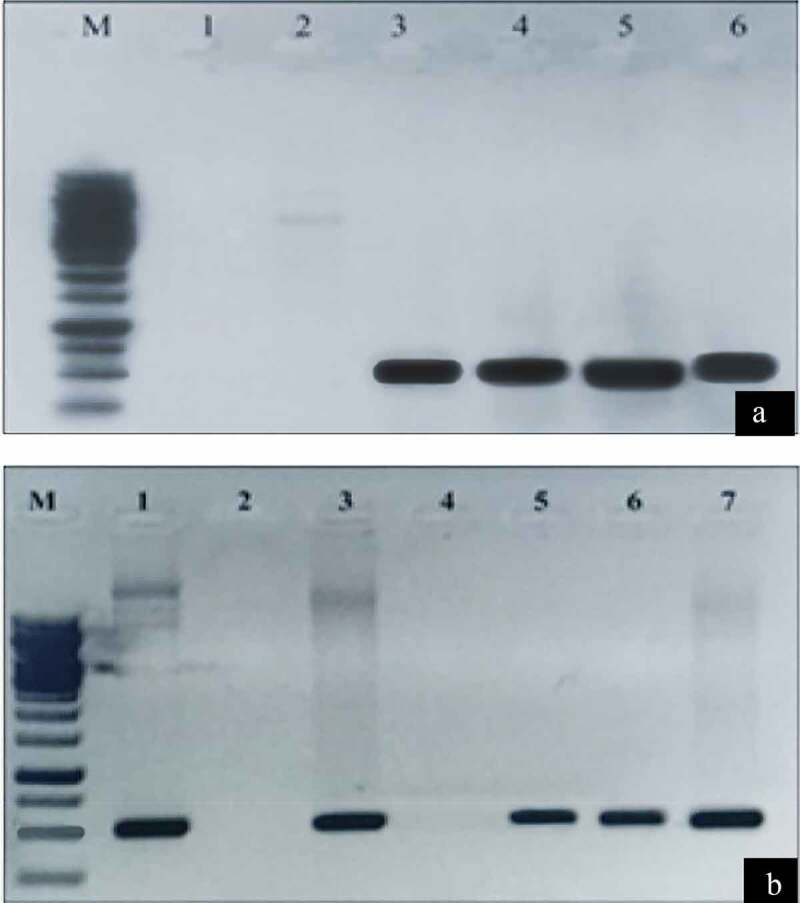
Determination of cry genes and PME genes expression in transgenic cotton plant by PCR analysis
Figure 4a, M: 10 Kb ladder; Lane 1, 2: shows negative sample; Lanes 3, 4: show positive for cry1Fa and Lane 5, 6: show positive for cry32Aa, positive samples. Figure 4b, M: 10 Kb ladder; Lane 1: shows positive control; Lane 2: shows negative control; Lanes 3, 5: show positive for AnPME, and Lane 6, 7: show positive for AtPME samples; lane 4: shows negative sample.
The mRNA from transgenic plants was isolated and cDNA transcribed. The relative mRNA expression of cry1Fa, cry32Aa, AtPME and AnPME genes was analyzed by using SYBR Green Mix in qPCR assays and higher expression of these genes was found in transgenic plants (Fig. 5). Expression of GAPDH gene was used as internal control reference for normalization in these assays. We analyzed the expression of these genes in four transgenic plants (named K1, K2, K3 and K4) that were positive for transformation with the four genes (cry1Fa, cry32A, AtPMe and AnPME). Control plants were non-transformed Eagle-2 cotton plants.
Figure 5.
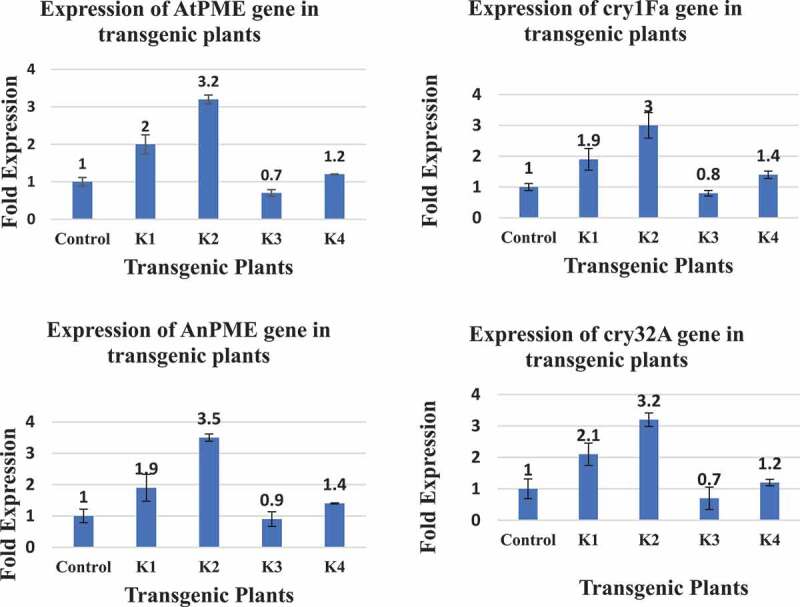
Relative expression of cry and PME genes in four transgenic cotton plants (K1, K2, K3 and K4)
The relative expression of cry and PME analyzed in different plants shown in the figure was calculated according to the 2 (-ΔΔCt) method using GAPDH as internal control reference gene for normalization.
The K2 plants showed the highest relative highest expression of the four AtPME and AnPME, cry1Fa and cry32A genes, while K3 plants showed the lower expression of these genes.
3.6. Evaluation of Methanol Concentration in Transgenic Cotton Plants
Transgenic K1-K4 cotton plants were subjected to Mass Spectrometry (MS) for methanol quantification. Transgenic plants K1 and K2 showed the highest contents of methanol, respectively, as compared to K3 and K4 plants and control plants. These data indicated that the methanol production in transgenic plants showed higher values than non-transgenic cotton plants (see Fig. 6).
Figure 6.
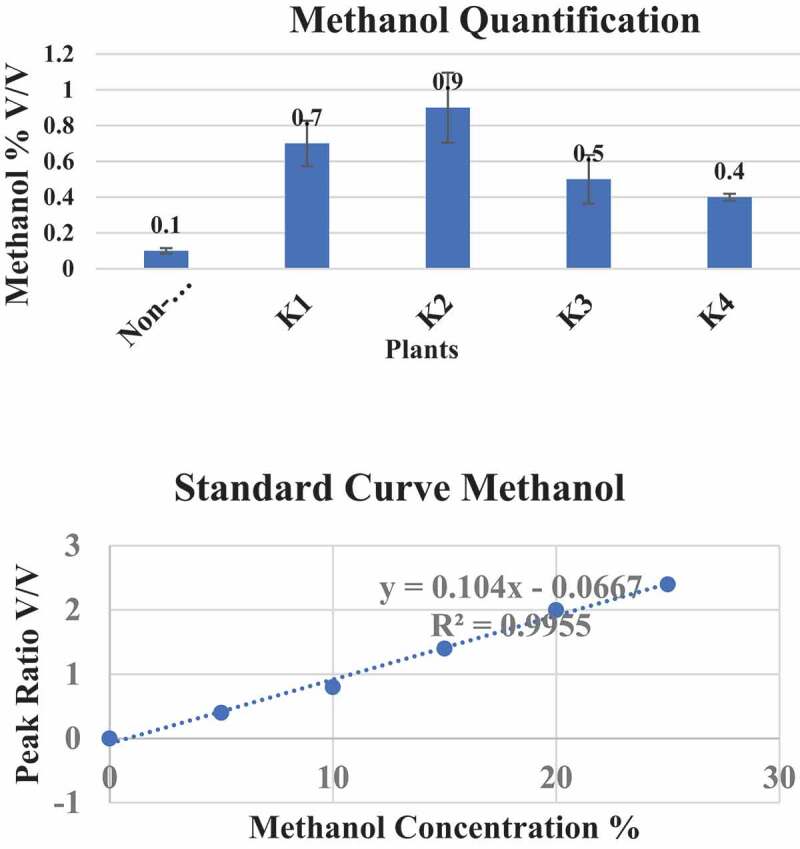
Methanol quantification in transgenic cotton plants. Standard curve of methanol was done by Mass Spectrometry (MS) with reference to a methanol standard. The transgenic cotton plants, namely, K1, K2, K3, K4 were subjected to methanol quantification
3.7. Evaluation of Transgenic Cotton Plants for Resistance against Chewing Insects
Bioassays were performed by using fresh leaves of non-transgenic cotton as negative control, and compared with transgenic cotton plants K1 to K4 expressing double Bt-cry genes and pme genes. We also compared with cotton plants transformed only with the two Bt cry-genes carried out in the laboratory. Each of the sampled fresh leaves was infected with H. armigera and P. gossypiella larvae. A 5–7 larvae of 3rd instar per leaf were used in triplicates.
The infestation H. armigera data were recorded, showing 100% mortality on the third day after infection of transgenic cotton plants harboring both Bt cry-genes coupled with AtPME and AnPME, while 84% mortality was observed after fifth days in the control plants transformed only with Bt-cry1Fa and cry32 genes and no mortality was observed in negative control of Eagle-2 cotton plants as shown in Fig. 7.
Figure 7.
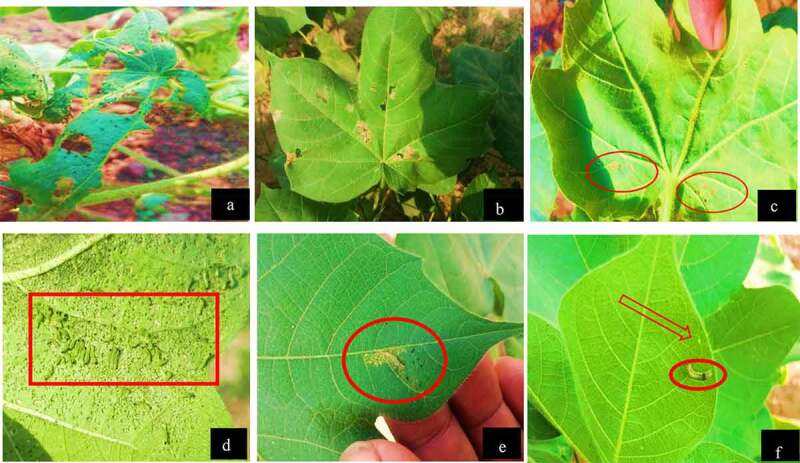
Cotton leaf chewing and mortality assay of H. armigera in different transgenic cotton plants
In the case of P. gossypiella larvae were released on freshly growing bolls of the cotton plants. Transgenic cotton plants expressing AtPME and AnPME along with Bt cry-genes showed 70% mortality of P. gossypiella larvae after 3 days, implying resistance toward P. gossypiella larvae. In contrast to transgenic cotton plants harboring only the two Bt cry-genes that showed 49% of P. gossypiella mortality and 0% mortality was observed in non-transgenic Eagle-2 control plants.
Finally, regarding spotted Bollworm (Earias fabia) 95% mortality was observed in transgenic harboring double Bt cry and pme gene cassettes, while 79% mortality was observed in transgenic plant expression only the double Bt cry-genes (Fig. 8). Two-way ANOVA showed the significance of our data at P≤ 0.0001 (Fig. 9).
Figure 8.

Pink Bollworm P. gossypiella mortality assays in different transgenic cotton plants. Figure 8a and b, in non-transgenic cotton plants, the cotton boll showed to be completely damaged by P. gossypiella and and larvae that were still alive are highlighted with a red circle mark. Figure 8c and d, in transgenic plants transformed with the double Bt-gene (cry1Fa and cry32Aa), the plant bolls showed insect resistance and only one locule was damaged but P. gossypiella larvae were dead. Figure 8e and f in the transgenic plants transformed with double Bt-gene (cry1Fa and cry32Aa) and double AnPME and AtPME gene in the same plant; the plants showed full resistance and even spotted was found dead on leaf and P. gossypiella free cotton was fully developed
Figure 9.
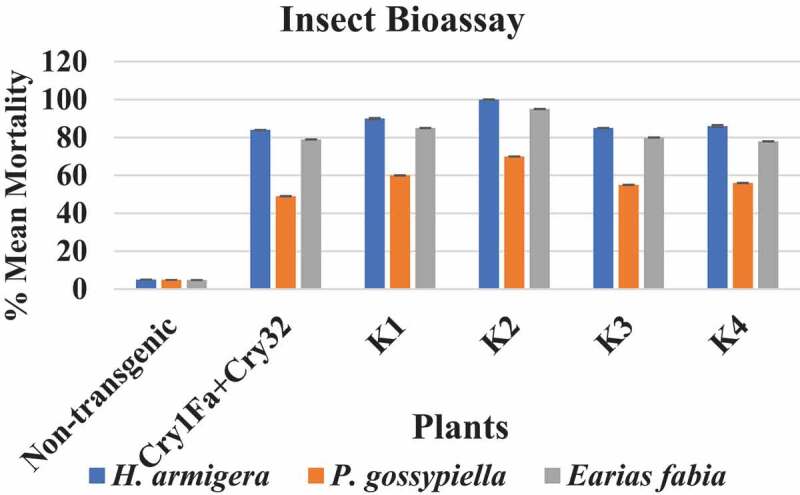
Mortality analysis against army boll worm Helicoverpa armigera, Pink boll worm Pectinophora gossypiella and spotted bollworm Earias fabia. Two-way ANOVA showed the significance of the data. compared to the non-transformed plants
Figure 7a, Non-transgenic cotton plant leaf completely chewed by H. armigera. Figure 7b, Double Bt-gene (cry1Fa and cry32Aa) plants showed resistance and a little bit chewed by H. armigera. Figure 7c, Double Bt-gene (cry1Fa and cry32Aa) and double AnPME and AtPME gene in the same plant showed full resistance and just a minor cut was observed by H. armigera. Figure 7d, Hatching of H. armigera on negative control Eagle-2 cotton plant leafs. Figure 7e, in double Bt-gene (cry1Fa and cry32Aa) cotton plant leafs, the H. armigera were dead after eating scratched looking portion of leaf. Figure 7f, In the double Bt-gene (cry1Fa and cry32Aa) and double AnPME and AtPME gene plants, the H. armigera were dead even after a minor first cut.
4. Discussion
Cotton is considered a socioeconomically important crop18–20 and due to current trend of global market it is necessary to make continuous improvements in cotton varieties.21 The transformation of new genes into cotton varieties is required to develop resistance against unrelated invading insect pests.22 For this goal, a unique approach was adopted therein two cassettes were designed. One cassette harbor two pectin methyl esterase genes (pme) from A. thaliana and As. niger and second cassette contains two insecticidal proteins coded by cry genes from B. thuringiensis. Both cassettes were transformed into non-transgenic cotton variety Eagle-2 to achieve resistance against insects as previously depicted in tobacco plants.8 Initial screening of putative cotton plants was done by hygromycin as reported.23 Putative transgenic cotton plants were confirmed through PCR by using specific primers as previously done.24 PCR confirmed transgenic plants were subjected to qPCR for determination of mRNA expression of the transgenic genes as reported.25 Ultimately, the efficacy of transgene was determined by insect bioassay against different target Lepidopteran larvae.26
Naturally, plants produce methanol, which is nontoxic for the plants.27 Methanol after accumulation in leaves is released into atmosphere through stomata28,29 and is found to be toxic to the insects.8 In this study, the methanol quantification was done in transgenic and non-transgenic plants by MS as reported before.10 Transgenic cotton plants K1 and K2 showed a 0.7% and 0.9% increase in methanol concentration when compared with the non-transgenic control plants, while K3 and K4 transformed plants showed only 0.5% and 0.4% methanol concentration, which is less than K1 and K2 plants, but greater than a control plant. Methanol concentration was calculated in correlation with insect bioassay in which K1 and K2 with higher expression of all genes showing 100% mortality in accordance with Hasunuma et al. (2003).
In this study, real-time qPCR assays were conducted to analyze mRNA expression of pme and cry genes in transgenic cotton plants. The relative expression of AtPME and AnPME was higher in K2 than in control plants. Insect bioassays were performed on detached leaves, flower and bolls of cotton. The infestation data were recorded and 100% mortality was observed after three days in transgenic cotton plants harboring both Bt cry-genes coupled with AtPME and AnPME genes, while 84% mortality was observed in Bt cry-genes transgenic cotton plants after five days, while there was no mortality observed in negative control cotton plants as shown in Fig. 8. The Pink Bollworm (P. gossypiella) larvae were released on freshly growing bolls of the cotton plants. Transgenic cotton plants showed resistance toward pink bollworm larvae for 3 days with 70% mortality in case of transgenic cotton plants harboring AtPME and AnPME along with Bt cry-genes in contrast with the 49% mortality observed in transgenic cotton plants harboring only the two Bt cry-genes and 0% mortality in non-transgenic control plants. Finally, we observed 95% mortality in spotted bollworm in transgenic plants harboring double Bt-genes and pme genes, while only 79% mortality was observed in transgenic plants expressing only the double Bt cry-genes as shown in Fig. 9. These results indicated that Bt cry-genes coupled with pme genes are a possible and useful strategy to control different insect pests and for lowering the resistance of insects against transgenic cotton varieties.
5. Conclusion
Transgenic plants expressing simultaneously pme and cry genes were evaluated against different Lepidopteran insect pests and compared against non-transgenic and transgenic plants expressing only Bt cry-genes. Increased mortality in insects was observed in transgenic plants harboring pme and cry gene combination as compared with positive control expressing cry genes only. The increased production of ethanol by pme genes in these transgenic plants explains their improvement in the control against insect attack. This control strategy infers that it may be robust to reduce the attack of different lethal cotton insects. As it is reported that insect attack has become a major concern in cotton growing countries around the world especially in Pakistan where farmers have started walking out from the cultivation of cotton due to the high risk of insect attack. Therefore, the proposed strategy may incur some positive results to win the farmer’s interest in the cotton cultivation.
6. Compliance with Ethical Standards
6.1. Potential Conflict Interest
The authors confirm that there is no actual or potential conflict of interest and also that the work described in this manuscript has not been published previously, that it is not under consideration for publication elsewhere. Its publication has been approved by all authors and implicitly by the responsible authorities where the work was carried out, and all persons entitled to authorship have been so named. The authors also confirm that, if accepted, it will not be published elsewhere including electronically in the same form, in English or in any other language, without the written consent of the copyright-holder.
6.2. Research Involvement with Human Participants/ or Animals
The research does not involve with human and animal participants.
6.3. Informed Consent
I have read and I understand the provided information and have granted the permission to ask the questions. I understand my participation in the research and liable to produce the given information at any stage.
9. Acknowledgements
I am grateful to The Institute of Cotton Research, Chinese Academy of Agricultural Sciences and FB Genetics Four Brothers Group on providing us the expert opinion and facilities performing the analysis and writing the entire manuscript.
7. Funding
This work was funded by the National Key R&D Program of China (2017YFD0101603-11, 2016YFD0100500), the Natural Science Foundation of China (31471538 and 31371668), the Agricultural Science and Technology Innovation Program for CAAS (CAAS-ASTIP-ICRCAAS), the National High Technology Research and Development Program of China (2012AA101108 and 2009AA101104) and the Central Level of the Scientific Research Institutes for Basic R & D Special Fund Business (1610162014008).
8. Authors’ contributions
AR, AA and MMZ wrote the initial draft of the manuscript and are equal contributors. AN, DX, MA and LP made all necessary corrections and carried out final editing of manuscript. WG, MR and GQ proof read the manuscript. Final approval for publication was given by the group leader at institute of cotton research YY.
Conflicts Of Interest
No potential conflict of interest was reported by the authors.
References
- 1.Tarazi R, JLS J, MF V.. Biotechnological solutions for major cotton (Gossypium hirsutum) pathogens and pests. Biotech Res and Innovation. 2020;3:19-26.. [Google Scholar]
- 2.Junyu L, Zhang S, Xiangzhen Z, Jichao J, Zhang K, Chunyi W, Zhang L, Li W, Jiniie C. Effects of NaCl stress on the biochemical substances in Bt cotton as well as on the growth and development and adult oviposition selectivity of Helicoverpa armigera. Jl of Cotton Res. 2019;2(1):4. doi: 10.1186/s42397-019-0020-7. [DOI] [Google Scholar]
- 3.Verma C, AKM T, Mishra S. Biochemical and molecular characterization of cell wall degrading enzyme, pectin methylesterase versus banana ripening: an overview. Asian J of Biotech. 2017;9(1):1–23. doi: 10.3923/ajbkr.2017.1.23. [DOI] [Google Scholar]
- 4.Williams M. Cotton insect losses. Mississippi State University; 1986-2015. 2016.
- 5.Gao M, Wang X, Yang Y, BE T, Wu Y. Epistasis confers resistance to Bt toxin Cry1Ac in the cotton bollworm. Evol Appl. 2018;11(5):809–19. doi: 10.1111/eva.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouser S, DJ S, Qaim M. Transgenic cotton and farmers’ health in Pakistan. PloS One. 2019;14(10):10. doi: 10.1371/journal.pone.0222617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.YL D, AV S, EV S, TV K. Metabolic methanol: molecular pathways and physiological roles. Physiol Rev. 2015;95(2):603–44. doi: 10.1152/physrev.00034.2014. [DOI] [PubMed] [Google Scholar]
- 8.Dixit S, SK U, Singh H, OP S, PC V, Chandrashekar K. Enhanced methanol production in plants provides broad spectrum insect resistance. PLoS One. 2013;8(11):e79664. doi: 10.1371/journal.pone.0079664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelloux J, Rusterucci C, EJ M. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007;12(6):267–77. doi: 10.1016/j.tplants.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Hasunuma T, E-i F, Kobayashi A. Methanol production is enhanced by expression of an Aspergillus niger pectin methylesterase in tobacco cells. J Biotechnol. 2003;106(1):45–52. doi: 10.1016/j.jbiotec.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Jirschitzka J, DJ M, Gershenzon J, JC D. Learning from nature: new approaches to the metabolic engineering of plant defense pathways. Curr Opin Biotechnol. 2013;24(2):320–28. doi: 10.1016/j.copbio.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Körner E, von D, Bonaventure G, IT B. Pectin methylesterase Na PME1 contributes to the emission of methanol during insect herbivory and to the elicitation of defence responses in Nicotiana attenuata. Journal of Experimental Botany. 2009;60(9):2631–40. doi: 10.1093/jxb/erp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopa L, Benny P. Selective comparison of repellent activity with contact toxicity of certain essential oils against different insect orders. Int J of Life Sciences. 2018;6:615–25. [Google Scholar]
- 14.Gopa L, PJIJoLS B. Selective comparison of repellent activity with contact toxicity of certain essential oils against different insect orders. Int J Life Sci. 2018;6(2):615-25. [Google Scholar]
- 15.Körner E, von D, Bonaventure G, ITJJoeb B. Pectin methylesterase Na PME1 contributes to the emission of methanol during insect herbivory and to the elicitation of defence responses in Nicotiana attenuata. Journal of Experimental Botany. 2009;60:2631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristina A, Maria Q, Vieira M-L. Plant transformation: advances and perspectives. Scientia Agricola. 1999;56(1):1-8. [Google Scholar]
- 17.Cubero J, Martinez M, Llop P, López M. A simple and efficient PCR method for the detection of Agrobacterium tumefaciens in plant tumours. J Appl Microbiol. 1999;86(4):591–602. doi: 10.1046/j.1365-2672.1999.00700.x. [DOI] [PubMed] [Google Scholar]
- 18.ABRAPA . Estatísticas: o Algodão no mundo. Associação Brasileira de Produtores de Algodão. 2018.
- 19..CONABA . companhamento da safra de grãos brasileira (SAFRA 2016/17 - Quarto levantamento). Companhia Nacional De Abastecimento 2018;4:1–162. [Google Scholar]
- 20.USDA . Cotton: world markets and trade 2017/18. Foreign agricultural Service/USDA. office of global analysis. 2018:1–27.
- 21.Trapero C, WilsonI W, WN S, LJ W. Enhancing integrated pest management in GM cotton systems using host plant resistance. Front Plant Sci. 2016;7:500. doi: 10.3389/fpls.2016.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrière Y, Crickmore N, BE T. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol. 2015;33(2):161. doi: 10.1038/nbt.3099. [DOI] [PubMed] [Google Scholar]
- 23.AK S, Pental D. Selection and genetic transformation of a fast-growing cell line in cotton (Gossypium hirsutum) for transgene expression studies. Jl of Plant Biochemistry and Biotech. 2015;24(2):225–32. doi: 10.1007/s13562-014-0262-x. [DOI] [Google Scholar]
- 24.Yasmeen A, Kiani S, Butt A, Rao AQ, Akram F, Ahmad A, Nasir IA, Husnain T, Mansoor S, Amin I, Aftab S. Amplicon-based RNA interference targeting V2 gene of cotton leaf curl Kokhran Virus-Burewala strain can provide resistance in transgenic cotton plants. Mol Biotechnol. 2016;58(12):807–20. doi: 10.1007/s12033-016-9980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao A, shahzad BK, PA N, UKM A, AM A, RM U, et al. Variation in expression of phytochrome B gene in cotton (Gossypium hirsutum L.). 2013 Sep 10;15(5):1033-42..
- 26.Khan A, NasirI A, Tabassum B, Aaliya K, Tariq M, AQ R. Expression studies of chitinase gene in transgenic potato against Alternaria solani. Plant Cell, Tissue and Organ Culture (PCTOC). 2017;128(3):563–76. doi: 10.1007/s11240-016-1134-y. [DOI] [Google Scholar]
- 27.Nemecek-Marshall M, RC M, JJ F, CL W, Fall R. Methanol emission from leaves (enzymatic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development). Plant Physiol. 1995;108(4):1359–68. doi: 10.1104/pp.108.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aslam M. Emission and role of biogenic volatile organic compounds in biosphere. A J of Sci for Development. 2014;19:69. [Google Scholar]
- 29.Hüve K, Christ MM, Kleist E, Uerlings R, Niinemets Ü, Walter A, Wildt J. Simultaneous growth and emission measurements demonstrate an interactive control of methanol release by leaf expansion and stomata. Journal of Experimental Botany. 2007;58(7):1783–93. doi: 10.1093/jxb/erm038. [DOI] [PubMed] [Google Scholar]


