Figure 6.
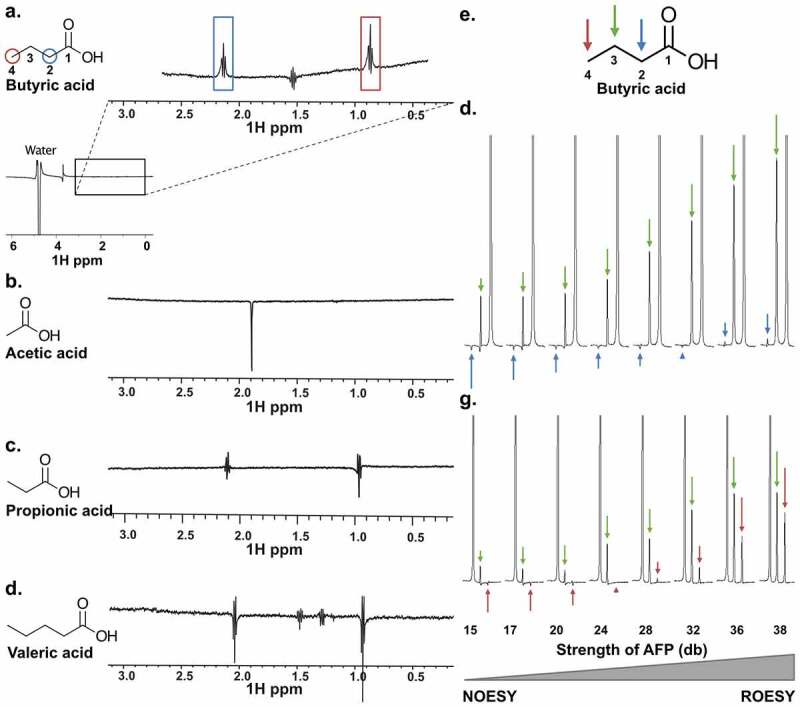
NMR characterization of SCFAs binding to PHD2181-402. (a-d) WaterLOGSY 1D NMR to determine SCFAs binding to PHD2181-402. 10 mM of each tested SCFA was mixed with 25 μM PHD2181-402. ((a) Butyric acid; peak inversions relative to the water signal were seen for protons directly bound to carbons C2 and C4, indicating binding. (b-d) Acetic acid, propionic acid and valeric acid, respectively; no peak is inverted, indicating no binding under these experimental conditions. (e-g) AFP-NOESY 1D NMR to determine butyrate atoms in the binding pocket of PHD2181-402. (e) The butyrate structure with labeled positions and corresponding arrow color code. (f) 10 mM butyrate was mixed with 25 μM PHD2181-402. The strength of the adiabatic pulse was gradually increased to shift relaxation contributions from longitudinal cross relaxation (NOESY) to transverse cross relaxation (ROESY). Protons attached to C4 were selectively inverted and acted as source of magnetization transfer. Peaks of protons attached to C2, unlike protons attached to C3, showed a profile typical for strong spin diffusion. This indicates embedding of the C2 in the binding pocket of the protein. (g) Protons attached to C2 were selectively inverted. Peaks of protons attached to C4, unlike protons attached to C3, showed a profile typical for strong spin diffusion, indicating embedding of the C4 protons in the binding pocket. The weak spin diffusion dependence of the protons attached to C3 in both (F) and (G) indicates that the C3 protons are more solvent exposed
