Figure 2.
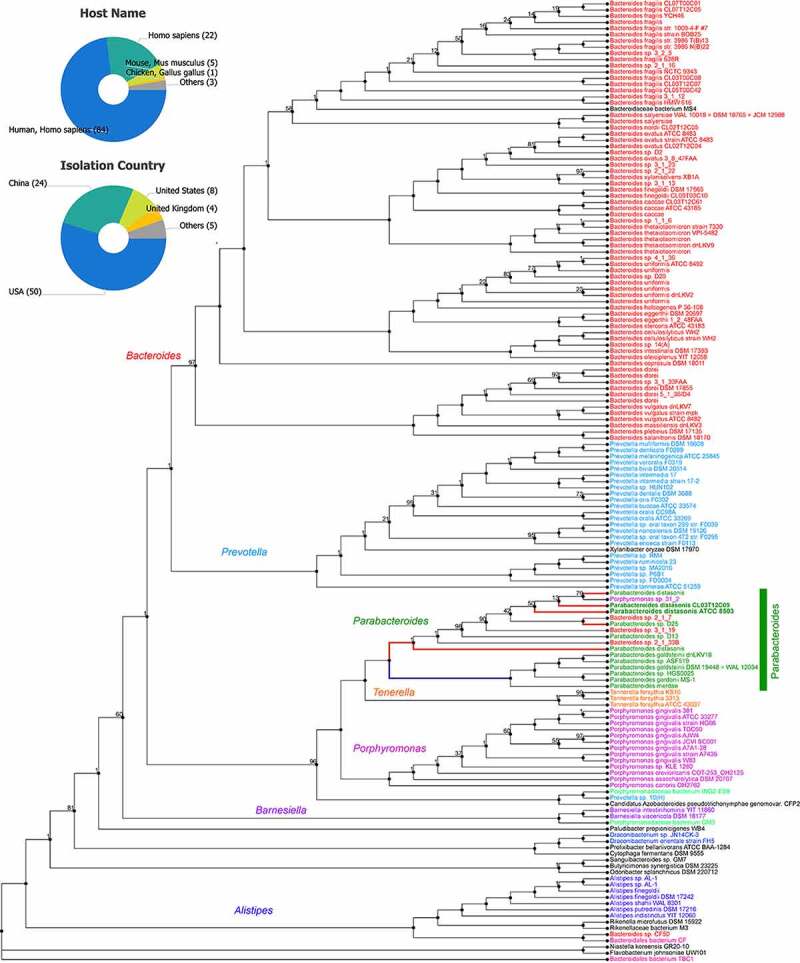
Protein phylogram of 155 complete genomes of the Bacteroidetes phylum to illustrate the potential functional distinction of Parabacteroides distasonis from other species within the genus. The pipeline for genomic phylograms is described in detail based on information from PATRIC, the Pathosystems Resource Integration Center, https://docs.patricbrc.org. In short, the order-level pre-built trees in PATRIC are constructed by an automated pipeline that begins with amino acid sequence files for each genome. For each order-level tree the genomes from that order are used along with a small set of potential outgroup genomes. Branch values are not bootstrap values, which can be overly optimistic for long genomes. Instead, trees are built from random samples of 50% of the homology groups used for the main tree (gene-wise jackknifing). One hundred of these 50% gene-wise jackknife trees are made using FastTree, and the support values shown indicate the number of times a particular branch was observed in the support trees. As of January 12, 2021, there were 133 P. distasonis genomes available, of which 8 are complete (pie charts).131
