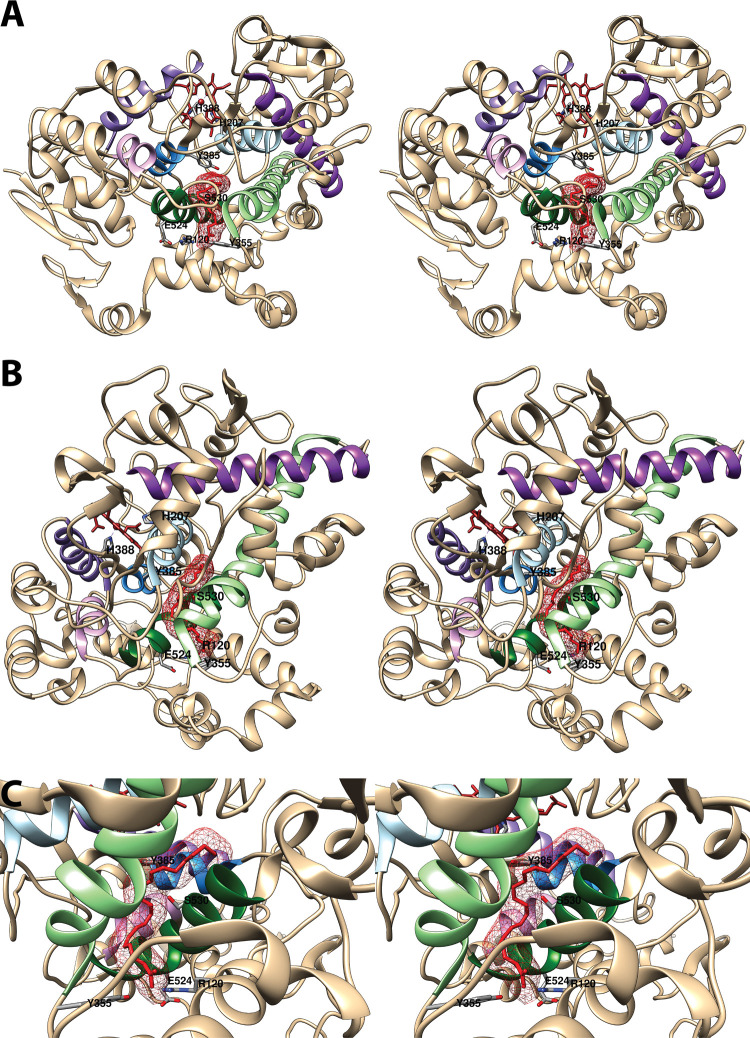
Figure 4.
Structure of the cyclooxygenase active site. Two wall-eyed stereo views of the COX-1 monomer (A and B) and a close-up view of the cyclooxygenase active site (C) are shown as observed from the side (i.e., parallel to the plane of the membrane). In all cases, Co3+-protoporphyrin IX (an inactive heme analog) is dark brown, and AA is red mesh. The side chains of the constriction residues (Arg-120, Tyr-355, and Glu-524) are displayed and labeled, as are the catalytic residue (Tyr-385) and the target of aspirin-mediated inactivation (Ser-530), which are located at the bend of the L-shaped channel. In A and B, His-388, the proximal heme ligand, and His-207, which serves as the distal heme ligand through a coordinating water molecule, are visible. Helices 2 (residues 195–207, light blue), 8 (residues 378–385, medium blue), 6 (residues 324–354, light green), and 17 (residues 519–535, dark green), which surround the active site, along with helices 5 (residues 295–320, dark purple), 11/12 (residues 444–459, medium purple), and 16 (residues 503–510, light purple) form a bundle that is conserved among a number of peroxidases, with helices 2, 5, 6, 8, and 11/12 involved in binding the heme prosthetic group. From PDB 1DIY.