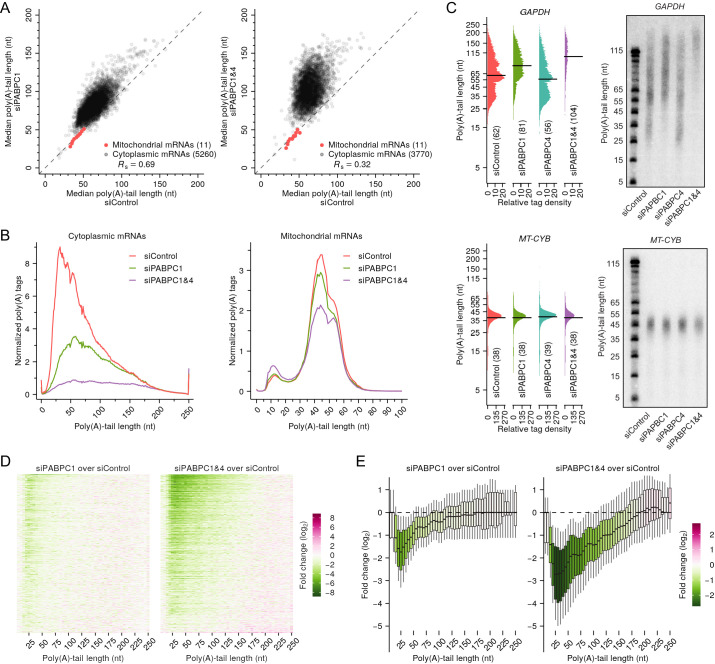
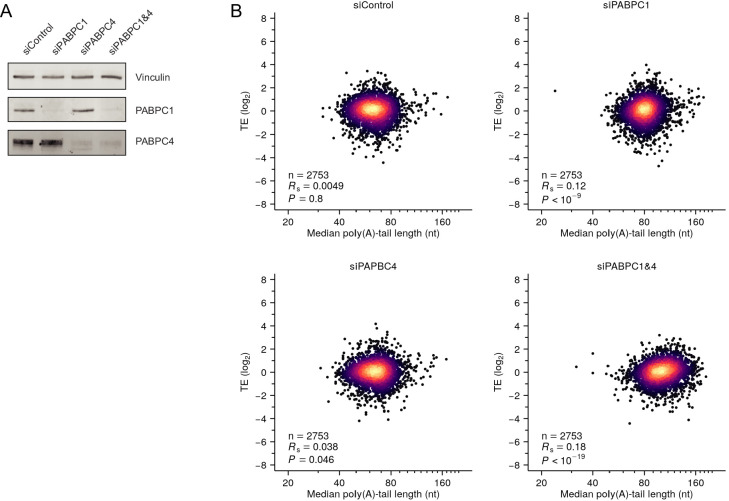
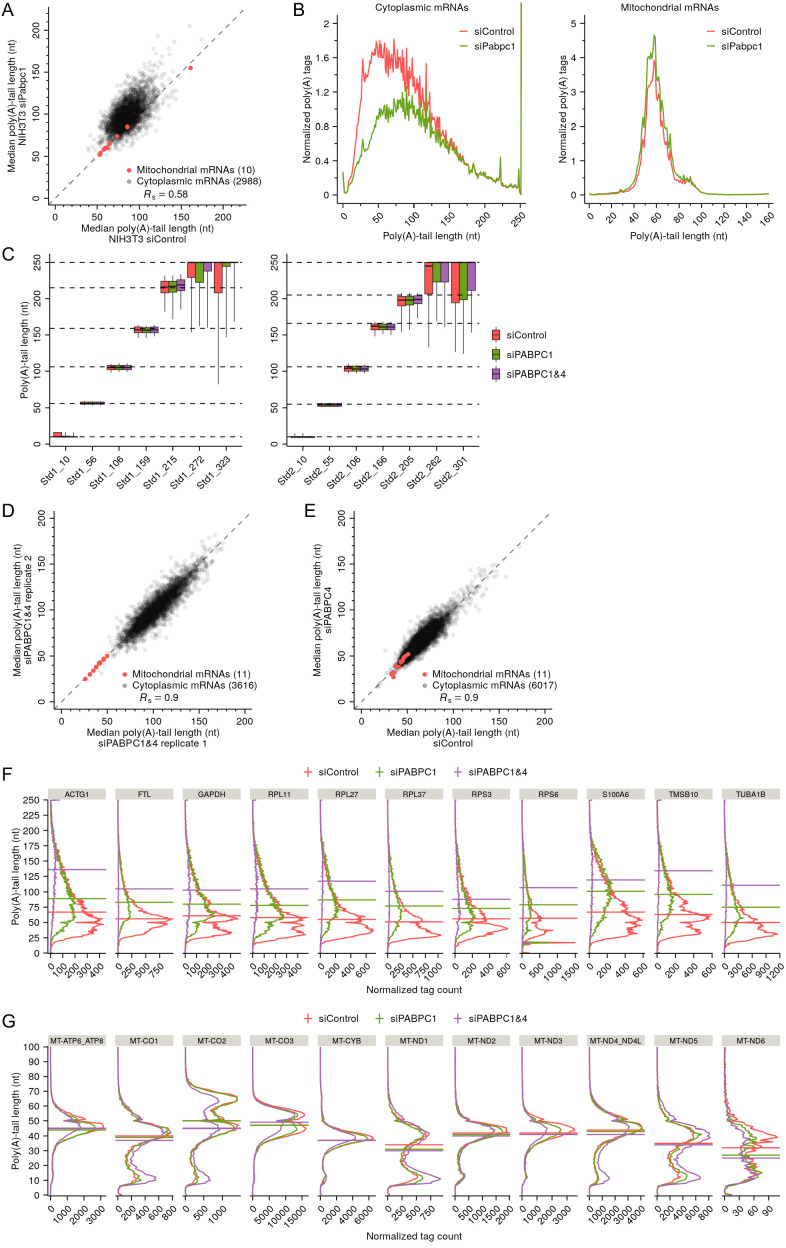
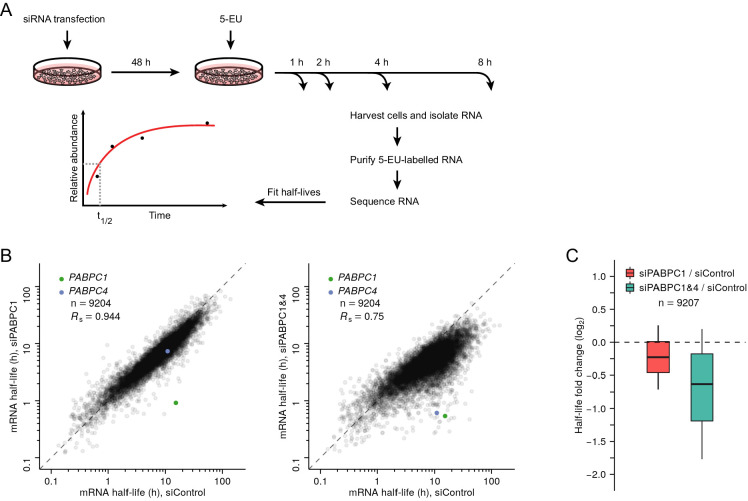
Figure 4. PABPC depletion causes premature decay of shorter-tail cytoplasmic mRNAs in HeLa cells.
(A) The effect of PABPC knockdown on poly(A)-tail length. The plots compare median poly(A)-tail lengths in either PABPC1-knockdown cells (left) or PABPC1 and PABPC4 double-knockdown cells (right) to those in control cells. Results are shown for cytoplasmic mRNAs with ≥100 poly(A) tags (gray) and for mitochondrial mRNAs (red), merging data for MT-ATP6 and MT-ATP8 and for MT-ND4 and MT-ND4L, which are bicistronic mitochondrial mRNAs. (B) The effect of PABPC knockdown on the abundance of mRNAs with different tail lengths. Shown are tail-length distributions of all cytoplasmic (left) and mitochondrial (right) mRNA poly(A) tags in control, PABPC1-knockdown, and double-knockdown cells. For each distribution, the abundance of tags was normalized to that of the spike-in tail-length standards. Due to depletion of tail-length calling at position 50, which was associated with a change in laser intensity at the next sequencing cycle, the values at this tail length were replaced with the average of values at tail lengths 49 and 51 nt. (C) The effect of PABPC knockdown on the abundance of mRNAs with different tail lengths, comparing tail-lengths measured by sequencing (left) with those observed on RNase H northern blots (right). Results are shown for a cytoplasmic mRNA GAPDH (top) and a mitochondrial mRNA MT-CYB (bottom). Relative tag density was calculated by log-transforming linear tag density using normalized poly(A) tag counts. Median tail-length values are indicated (horizontal lines) and listed in parentheses. For RNase H northern blots, a DNA oligonucleotide complimentary to the 3′-UTR was used to direct cleavage of the target mRNA by RNase H, leaving a 35-nt fragment of the 3′-UTR appended to the poly(A) tail, which was resolved on a denaturing gel and detected by a radiolabeled probe. Tail lengths indicated along the left side of each gel are inferred from lengths of size markers. (D) The effect of PABPC knockdown on the abundance of mRNAs with different tail lengths, extending the intragenic analysis to tail-length distributions from thousands of genes. Heat maps compare poly(A)-tag levels in PABPC1-knockdown (left) or double-knockdown (right) cells to those in control cells, after normalizing to spike-in tail-length standards, as measured using tail-length sequencing. Each row represents mRNAs from a different gene, and rows are sorted based on fold change of mRNA abundance measured using RNA-seq. Only genes with ≥100 poly(A) tags in each of two samples being compared were included in the analyses (n = 5504). Columns represent values from 5-nt tail-length bins ranging from 0 to 244 nt and a 6-nt bin ranging from 245 to 250 nt. Tile color indicates the fold change of normalized tag counts (key). (E) The effect of PABPC knockdown on the abundance of mRNAs with different tail lengths, reanalyzing data from (D) to show distributions of poly(A)-tag changes observed at different tail-lengths. Each box-whisker shows the 10th, 25th, 50th, 75th, and 90th percentile of fold changes in normalized poly(A)-tag counts observed for each tail-length bin of (D). The color of each box indicates the median value (key). Tail-length measurements in this figure were obtained using TAIL-seq.